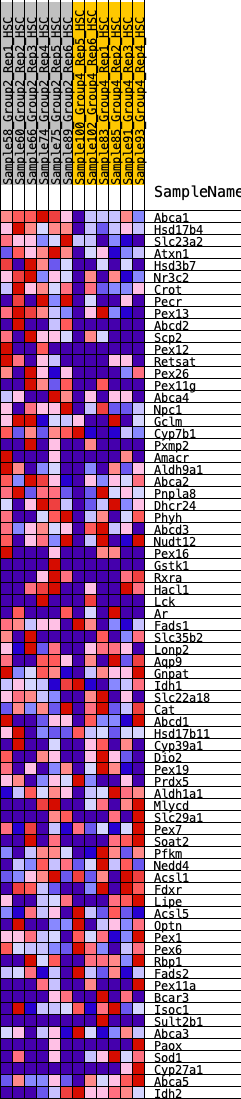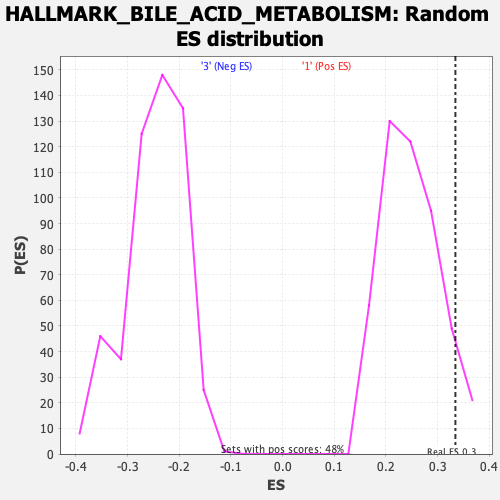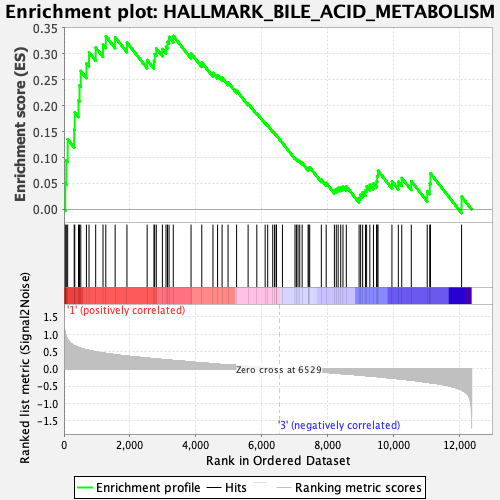
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.33453694 |
| Normalized Enrichment Score (NES) | 1.3546426 |
| Nominal p-value | 0.09052631 |
| FDR q-value | 0.75145423 |
| FWER p-Value | 0.58 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 39 | 1.009 | 0.0495 | Yes |
| 2 | Hsd17b4 | 70 | 0.910 | 0.0946 | Yes |
| 3 | Slc23a2 | 111 | 0.839 | 0.1351 | Yes |
| 4 | Atxn1 | 309 | 0.669 | 0.1540 | Yes |
| 5 | Hsd3b7 | 327 | 0.662 | 0.1872 | Yes |
| 6 | Nr3c2 | 440 | 0.614 | 0.2102 | Yes |
| 7 | Crot | 471 | 0.602 | 0.2392 | Yes |
| 8 | Pecr | 509 | 0.590 | 0.2670 | Yes |
| 9 | Pex13 | 680 | 0.544 | 0.2815 | Yes |
| 10 | Abcd2 | 759 | 0.530 | 0.3029 | Yes |
| 11 | Scp2 | 961 | 0.488 | 0.3120 | Yes |
| 12 | Pex12 | 1183 | 0.451 | 0.3175 | Yes |
| 13 | Retsat | 1268 | 0.441 | 0.3337 | Yes |
| 14 | Pex26 | 1551 | 0.404 | 0.3319 | Yes |
| 15 | Pex11g | 1909 | 0.363 | 0.3218 | Yes |
| 16 | Abca4 | 2522 | 0.307 | 0.2880 | Yes |
| 17 | Npc1 | 2728 | 0.289 | 0.2864 | Yes |
| 18 | Gclm | 2751 | 0.285 | 0.2995 | Yes |
| 19 | Cyp7b1 | 2799 | 0.281 | 0.3103 | Yes |
| 20 | Pxmp2 | 2984 | 0.265 | 0.3092 | Yes |
| 21 | Amacr | 3094 | 0.255 | 0.3137 | Yes |
| 22 | Aldh9a1 | 3139 | 0.252 | 0.3232 | Yes |
| 23 | Abca2 | 3186 | 0.248 | 0.3324 | Yes |
| 24 | Pnpla8 | 3314 | 0.238 | 0.3345 | Yes |
| 25 | Dhcr24 | 3853 | 0.195 | 0.3009 | No |
| 26 | Phyh | 4179 | 0.169 | 0.2833 | No |
| 27 | Abcd3 | 4518 | 0.143 | 0.2633 | No |
| 28 | Nudt12 | 4658 | 0.131 | 0.2589 | No |
| 29 | Pex16 | 4795 | 0.121 | 0.2541 | No |
| 30 | Gstk1 | 4976 | 0.110 | 0.2452 | No |
| 31 | Rxra | 5235 | 0.094 | 0.2291 | No |
| 32 | Hacl1 | 5584 | 0.068 | 0.2044 | No |
| 33 | Lck | 5848 | 0.050 | 0.1856 | No |
| 34 | Ar | 6101 | 0.032 | 0.1668 | No |
| 35 | Fads1 | 6178 | 0.026 | 0.1619 | No |
| 36 | Slc35b2 | 6331 | 0.015 | 0.1503 | No |
| 37 | Lonp2 | 6387 | 0.010 | 0.1464 | No |
| 38 | Aqp9 | 6434 | 0.007 | 0.1430 | No |
| 39 | Gnpat | 6444 | 0.006 | 0.1426 | No |
| 40 | Idh1 | 6625 | -0.004 | 0.1281 | No |
| 41 | Slc22a18 | 7006 | -0.030 | 0.0988 | No |
| 42 | Cat | 7049 | -0.033 | 0.0970 | No |
| 43 | Abcd1 | 7097 | -0.036 | 0.0951 | No |
| 44 | Hsd17b11 | 7150 | -0.041 | 0.0930 | No |
| 45 | Cyp39a1 | 7223 | -0.047 | 0.0896 | No |
| 46 | Dio2 | 7403 | -0.062 | 0.0783 | No |
| 47 | Pex19 | 7427 | -0.065 | 0.0798 | No |
| 48 | Prdx5 | 7452 | -0.066 | 0.0813 | No |
| 49 | Aldh1a1 | 7805 | -0.093 | 0.0575 | No |
| 50 | Mlycd | 7952 | -0.103 | 0.0510 | No |
| 51 | Slc29a1 | 8201 | -0.122 | 0.0372 | No |
| 52 | Pex7 | 8258 | -0.127 | 0.0392 | No |
| 53 | Soat2 | 8315 | -0.132 | 0.0416 | No |
| 54 | Pfkm | 8393 | -0.138 | 0.0425 | No |
| 55 | Nedd4 | 8461 | -0.143 | 0.0445 | No |
| 56 | Acsl1 | 8562 | -0.152 | 0.0443 | No |
| 57 | Fdxr | 8950 | -0.181 | 0.0223 | No |
| 58 | Lipe | 8997 | -0.186 | 0.0282 | No |
| 59 | Acsl5 | 9058 | -0.188 | 0.0332 | No |
| 60 | Optn | 9143 | -0.197 | 0.0367 | No |
| 61 | Pex1 | 9178 | -0.201 | 0.0444 | No |
| 62 | Pex6 | 9276 | -0.211 | 0.0475 | No |
| 63 | Rbp1 | 9385 | -0.217 | 0.0501 | No |
| 64 | Fads2 | 9480 | -0.225 | 0.0542 | No |
| 65 | Pex11a | 9499 | -0.226 | 0.0645 | No |
| 66 | Bcar3 | 9524 | -0.229 | 0.0745 | No |
| 67 | Isoc1 | 9946 | -0.269 | 0.0543 | No |
| 68 | Sult2b1 | 10140 | -0.287 | 0.0536 | No |
| 69 | Abca3 | 10244 | -0.297 | 0.0607 | No |
| 70 | Paox | 10533 | -0.328 | 0.0544 | No |
| 71 | Sod1 | 11016 | -0.388 | 0.0354 | No |
| 72 | Cyp27a1 | 11093 | -0.401 | 0.0502 | No |
| 73 | Abca5 | 11115 | -0.403 | 0.0695 | No |
| 74 | Idh2 | 12059 | -0.612 | 0.0247 | No |