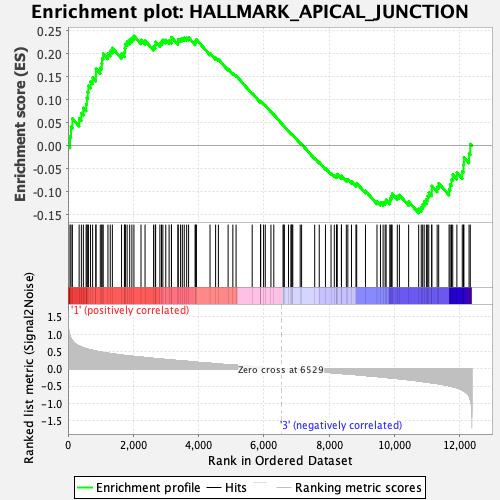
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group4.HSC_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.2391371 |
| Normalized Enrichment Score (NES) | 1.0277308 |
| Nominal p-value | 0.4108527 |
| FDR q-value | 0.68486637 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nectin1 | 61 | 0.943 | 0.0201 | Yes |
| 2 | Crat | 92 | 0.878 | 0.0411 | Yes |
| 3 | Amigo2 | 134 | 0.809 | 0.0593 | Yes |
| 4 | Lima1 | 341 | 0.656 | 0.0599 | Yes |
| 5 | Syk | 409 | 0.629 | 0.0712 | Yes |
| 6 | Cdk8 | 470 | 0.602 | 0.0824 | Yes |
| 7 | Pik3cb | 555 | 0.578 | 0.0909 | Yes |
| 8 | Adam15 | 578 | 0.569 | 0.1042 | Yes |
| 9 | Ikbkg | 605 | 0.563 | 0.1171 | Yes |
| 10 | Ptprc | 625 | 0.556 | 0.1304 | Yes |
| 11 | Pkd1 | 688 | 0.543 | 0.1398 | Yes |
| 12 | Actg1 | 754 | 0.530 | 0.1486 | Yes |
| 13 | Rsu1 | 854 | 0.510 | 0.1541 | Yes |
| 14 | Nectin3 | 855 | 0.509 | 0.1676 | Yes |
| 15 | Actn1 | 988 | 0.485 | 0.1698 | Yes |
| 16 | Sirpa | 1033 | 0.476 | 0.1789 | Yes |
| 17 | Myl12b | 1044 | 0.475 | 0.1907 | Yes |
| 18 | Pten | 1078 | 0.470 | 0.2005 | Yes |
| 19 | Nectin4 | 1223 | 0.446 | 0.2006 | Yes |
| 20 | Tjp1 | 1293 | 0.438 | 0.2067 | Yes |
| 21 | Plcg1 | 1358 | 0.429 | 0.2129 | Yes |
| 22 | Pard6g | 1639 | 0.392 | 0.2004 | Yes |
| 23 | Cd86 | 1727 | 0.382 | 0.2035 | Yes |
| 24 | Src | 1742 | 0.380 | 0.2125 | Yes |
| 25 | Dlg1 | 1756 | 0.379 | 0.2215 | Yes |
| 26 | Myh9 | 1810 | 0.374 | 0.2272 | Yes |
| 27 | Map4k2 | 1889 | 0.365 | 0.2305 | Yes |
| 28 | Nrap | 1956 | 0.359 | 0.2347 | Yes |
| 29 | Cap1 | 2018 | 0.353 | 0.2391 | Yes |
| 30 | Lama3 | 2235 | 0.332 | 0.2303 | No |
| 31 | Sgce | 2359 | 0.321 | 0.2288 | No |
| 32 | Nrtn | 2620 | 0.296 | 0.2155 | No |
| 33 | Ctnna1 | 2670 | 0.294 | 0.2193 | No |
| 34 | Itga9 | 2683 | 0.293 | 0.2261 | No |
| 35 | Speg | 2813 | 0.280 | 0.2230 | No |
| 36 | Cnn2 | 2862 | 0.277 | 0.2265 | No |
| 37 | Gnai1 | 2904 | 0.272 | 0.2304 | No |
| 38 | Epb41l2 | 2993 | 0.265 | 0.2303 | No |
| 39 | Tro | 3095 | 0.255 | 0.2288 | No |
| 40 | Slit2 | 3166 | 0.249 | 0.2297 | No |
| 41 | Lamb3 | 3168 | 0.249 | 0.2363 | No |
| 42 | Wasl | 3365 | 0.233 | 0.2265 | No |
| 43 | Tspan4 | 3374 | 0.232 | 0.2320 | No |
| 44 | Cd34 | 3447 | 0.227 | 0.2322 | No |
| 45 | Skap2 | 3500 | 0.222 | 0.2339 | No |
| 46 | Adam9 | 3559 | 0.218 | 0.2349 | No |
| 47 | Baiap2 | 3633 | 0.214 | 0.2347 | No |
| 48 | Evl | 3692 | 0.209 | 0.2355 | No |
| 49 | Mpzl1 | 3891 | 0.191 | 0.2244 | No |
| 50 | Dhx16 | 3908 | 0.189 | 0.2281 | No |
| 51 | Pals1 | 3932 | 0.187 | 0.2312 | No |
| 52 | Gnai2 | 4348 | 0.156 | 0.2015 | No |
| 53 | Fscn1 | 4519 | 0.143 | 0.1914 | No |
| 54 | Akt2 | 4604 | 0.136 | 0.1882 | No |
| 55 | Icam2 | 4902 | 0.115 | 0.1670 | No |
| 56 | Vcl | 5047 | 0.108 | 0.1581 | No |
| 57 | Myh10 | 5146 | 0.102 | 0.1528 | No |
| 58 | Arhgef6 | 5639 | 0.065 | 0.1143 | No |
| 59 | Mmp2 | 5894 | 0.047 | 0.0948 | No |
| 60 | Exoc4 | 5895 | 0.047 | 0.0961 | No |
| 61 | Jup | 5898 | 0.047 | 0.0972 | No |
| 62 | Rhof | 5988 | 0.040 | 0.0910 | No |
| 63 | Tsc1 | 6040 | 0.036 | 0.0878 | No |
| 64 | Tial1 | 6213 | 0.023 | 0.0743 | No |
| 65 | Zyx | 6301 | 0.016 | 0.0677 | No |
| 66 | Msn | 6583 | -0.001 | 0.0447 | No |
| 67 | Pbx2 | 6606 | -0.002 | 0.0430 | No |
| 68 | Shc1 | 6623 | -0.004 | 0.0418 | No |
| 69 | Cercam | 6751 | -0.013 | 0.0318 | No |
| 70 | Bmp1 | 6824 | -0.018 | 0.0264 | No |
| 71 | Vwf | 6851 | -0.019 | 0.0247 | No |
| 72 | Mapk14 | 6859 | -0.020 | 0.0247 | No |
| 73 | Itga3 | 6882 | -0.022 | 0.0235 | No |
| 74 | Itga2 | 7109 | -0.038 | 0.0060 | No |
| 75 | Cadm2 | 7152 | -0.041 | 0.0037 | No |
| 76 | Nrxn2 | 7553 | -0.074 | -0.0270 | No |
| 77 | Jam3 | 7692 | -0.085 | -0.0360 | No |
| 78 | Arpc2 | 7882 | -0.098 | -0.0489 | No |
| 79 | Hadh | 8053 | -0.111 | -0.0598 | No |
| 80 | Map3k20 | 8153 | -0.117 | -0.0648 | No |
| 81 | Itgb1 | 8219 | -0.123 | -0.0668 | No |
| 82 | Pecam1 | 8232 | -0.124 | -0.0645 | No |
| 83 | Ptk2 | 8239 | -0.125 | -0.0616 | No |
| 84 | Vav2 | 8370 | -0.136 | -0.0686 | No |
| 85 | Cntn1 | 8373 | -0.136 | -0.0652 | No |
| 86 | Gtf2f1 | 8521 | -0.149 | -0.0732 | No |
| 87 | Nlgn2 | 8567 | -0.152 | -0.0728 | No |
| 88 | Actn4 | 8679 | -0.160 | -0.0776 | No |
| 89 | Sorbs3 | 8816 | -0.169 | -0.0842 | No |
| 90 | Taok2 | 8839 | -0.171 | -0.0815 | No |
| 91 | Layn | 9107 | -0.193 | -0.0982 | No |
| 92 | Cd274 | 9460 | -0.223 | -0.1210 | No |
| 93 | Ldlrap1 | 9566 | -0.233 | -0.1233 | No |
| 94 | Sdc3 | 9647 | -0.242 | -0.1234 | No |
| 95 | Tgfbi | 9709 | -0.248 | -0.1218 | No |
| 96 | Sympk | 9739 | -0.251 | -0.1175 | No |
| 97 | Nf2 | 9856 | -0.261 | -0.1200 | No |
| 98 | Actb | 9865 | -0.262 | -0.1137 | No |
| 99 | Col16a1 | 9901 | -0.263 | -0.1096 | No |
| 100 | B4galt1 | 9923 | -0.266 | -0.1042 | No |
| 101 | Vcan | 10082 | -0.282 | -0.1096 | No |
| 102 | Cx3cl1 | 10146 | -0.287 | -0.1071 | No |
| 103 | Ctnnd1 | 10429 | -0.316 | -0.1217 | No |
| 104 | Ywhah | 10736 | -0.352 | -0.1373 | No |
| 105 | Shroom2 | 10815 | -0.362 | -0.1341 | No |
| 106 | Rac2 | 10859 | -0.369 | -0.1278 | No |
| 107 | Vasp | 10905 | -0.375 | -0.1215 | No |
| 108 | Icam1 | 10973 | -0.384 | -0.1167 | No |
| 109 | Itga10 | 11012 | -0.388 | -0.1095 | No |
| 110 | Kcnh2 | 11046 | -0.393 | -0.1017 | No |
| 111 | Tubg1 | 11136 | -0.405 | -0.0982 | No |
| 112 | Cd276 | 11137 | -0.405 | -0.0874 | No |
| 113 | Insig1 | 11302 | -0.427 | -0.0894 | No |
| 114 | Nf1 | 11349 | -0.432 | -0.0817 | No |
| 115 | Rras | 11667 | -0.489 | -0.0946 | No |
| 116 | Akt3 | 11700 | -0.501 | -0.0838 | No |
| 117 | Rasa1 | 11741 | -0.508 | -0.0736 | No |
| 118 | Traf1 | 11779 | -0.518 | -0.0628 | No |
| 119 | Fbn1 | 11906 | -0.552 | -0.0584 | No |
| 120 | Stx4a | 12071 | -0.618 | -0.0553 | No |
| 121 | Pfn1 | 12109 | -0.642 | -0.0412 | No |
| 122 | Tmem8b | 12121 | -0.645 | -0.0249 | No |
| 123 | Inppl1 | 12279 | -0.778 | -0.0170 | No |
| 124 | Amigo1 | 12316 | -0.894 | 0.0038 | No |

