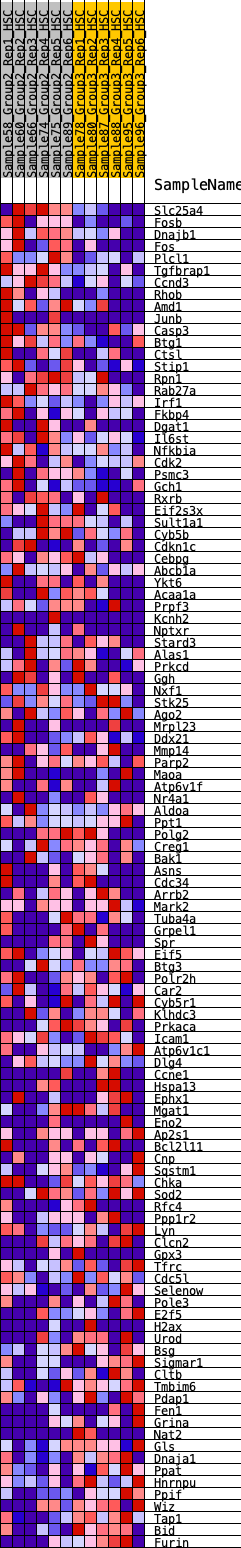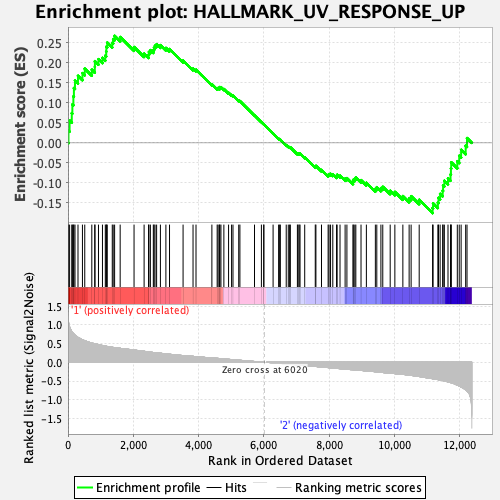
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.26764947 |
| Normalized Enrichment Score (NES) | 1.1256356 |
| Nominal p-value | 0.26611227 |
| FDR q-value | 0.49026257 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc25a4 | 15 | 1.084 | 0.0305 | Yes |
| 2 | Fosb | 49 | 0.955 | 0.0558 | Yes |
| 3 | Dnajb1 | 116 | 0.818 | 0.0743 | Yes |
| 4 | Fos | 134 | 0.795 | 0.0962 | Yes |
| 5 | Plcl1 | 165 | 0.769 | 0.1163 | Yes |
| 6 | Tgfbrap1 | 181 | 0.755 | 0.1372 | Yes |
| 7 | Ccnd3 | 214 | 0.728 | 0.1559 | Yes |
| 8 | Rhob | 306 | 0.661 | 0.1678 | Yes |
| 9 | Amd1 | 442 | 0.603 | 0.1744 | Yes |
| 10 | Junb | 513 | 0.576 | 0.1856 | Yes |
| 11 | Casp3 | 729 | 0.512 | 0.1830 | Yes |
| 12 | Btg1 | 821 | 0.494 | 0.1901 | Yes |
| 13 | Ctsl | 824 | 0.494 | 0.2044 | Yes |
| 14 | Stip1 | 933 | 0.471 | 0.2093 | Yes |
| 15 | Rpn1 | 1055 | 0.449 | 0.2126 | Yes |
| 16 | Rab27a | 1142 | 0.433 | 0.2183 | Yes |
| 17 | Irf1 | 1170 | 0.427 | 0.2286 | Yes |
| 18 | Fkbp4 | 1175 | 0.426 | 0.2407 | Yes |
| 19 | Dgat1 | 1202 | 0.423 | 0.2510 | Yes |
| 20 | Il6st | 1355 | 0.400 | 0.2503 | Yes |
| 21 | Nfkbia | 1383 | 0.397 | 0.2597 | Yes |
| 22 | Cdk2 | 1427 | 0.392 | 0.2676 | Yes |
| 23 | Psmc3 | 1597 | 0.373 | 0.2648 | No |
| 24 | Gch1 | 2022 | 0.328 | 0.2398 | No |
| 25 | Rxrb | 2330 | 0.294 | 0.2233 | No |
| 26 | Eif2s3x | 2466 | 0.278 | 0.2204 | No |
| 27 | Sult1a1 | 2478 | 0.276 | 0.2276 | No |
| 28 | Cyb5b | 2522 | 0.269 | 0.2320 | No |
| 29 | Cdkn1c | 2612 | 0.261 | 0.2324 | No |
| 30 | Cebpg | 2634 | 0.259 | 0.2382 | No |
| 31 | Abcb1a | 2664 | 0.256 | 0.2433 | No |
| 32 | Ykt6 | 2713 | 0.252 | 0.2468 | No |
| 33 | Acaa1a | 2829 | 0.242 | 0.2445 | No |
| 34 | Prpf3 | 2997 | 0.225 | 0.2374 | No |
| 35 | Kcnh2 | 3108 | 0.215 | 0.2348 | No |
| 36 | Nptxr | 3523 | 0.178 | 0.2062 | No |
| 37 | Stard3 | 3828 | 0.158 | 0.1860 | No |
| 38 | Alas1 | 3919 | 0.151 | 0.1831 | No |
| 39 | Prkcd | 4405 | 0.113 | 0.1468 | No |
| 40 | Ggh | 4567 | 0.105 | 0.1367 | No |
| 41 | Nxf1 | 4597 | 0.103 | 0.1373 | No |
| 42 | Stk25 | 4643 | 0.099 | 0.1366 | No |
| 43 | Ago2 | 4644 | 0.099 | 0.1395 | No |
| 44 | Mrpl23 | 4682 | 0.097 | 0.1393 | No |
| 45 | Ddx21 | 4772 | 0.089 | 0.1347 | No |
| 46 | Mmp14 | 4916 | 0.078 | 0.1253 | No |
| 47 | Parp2 | 5008 | 0.072 | 0.1199 | No |
| 48 | Maoa | 5051 | 0.069 | 0.1185 | No |
| 49 | Atp6v1f | 5227 | 0.056 | 0.1059 | No |
| 50 | Nr4a1 | 5263 | 0.053 | 0.1046 | No |
| 51 | Aldoa | 5710 | 0.021 | 0.0688 | No |
| 52 | Ppt1 | 5921 | 0.008 | 0.0519 | No |
| 53 | Polg2 | 5992 | 0.002 | 0.0462 | No |
| 54 | Creg1 | 5997 | 0.002 | 0.0460 | No |
| 55 | Bak1 | 6279 | -0.013 | 0.0234 | No |
| 56 | Asns | 6448 | -0.025 | 0.0104 | No |
| 57 | Cdc34 | 6479 | -0.028 | 0.0088 | No |
| 58 | Arrb2 | 6501 | -0.029 | 0.0079 | No |
| 59 | Mark2 | 6681 | -0.043 | -0.0054 | No |
| 60 | Tuba4a | 6744 | -0.047 | -0.0091 | No |
| 61 | Grpel1 | 6785 | -0.049 | -0.0109 | No |
| 62 | Spr | 6807 | -0.051 | -0.0112 | No |
| 63 | Eif5 | 7023 | -0.067 | -0.0267 | No |
| 64 | Btg3 | 7045 | -0.069 | -0.0265 | No |
| 65 | Polr2h | 7078 | -0.070 | -0.0270 | No |
| 66 | Car2 | 7104 | -0.072 | -0.0269 | No |
| 67 | Cyb5r1 | 7247 | -0.084 | -0.0361 | No |
| 68 | Klhdc3 | 7570 | -0.110 | -0.0591 | No |
| 69 | Prkaca | 7590 | -0.111 | -0.0574 | No |
| 70 | Icam1 | 7761 | -0.125 | -0.0677 | No |
| 71 | Atp6v1c1 | 7964 | -0.140 | -0.0800 | No |
| 72 | Dlg4 | 7992 | -0.142 | -0.0781 | No |
| 73 | Ccne1 | 8038 | -0.147 | -0.0775 | No |
| 74 | Hspa13 | 8107 | -0.152 | -0.0785 | No |
| 75 | Ephx1 | 8222 | -0.161 | -0.0831 | No |
| 76 | Mgat1 | 8239 | -0.162 | -0.0797 | No |
| 77 | Eno2 | 8328 | -0.169 | -0.0819 | No |
| 78 | Ap2s1 | 8484 | -0.182 | -0.0892 | No |
| 79 | Bcl2l11 | 8539 | -0.185 | -0.0882 | No |
| 80 | Cnp | 8727 | -0.197 | -0.0977 | No |
| 81 | Sqstm1 | 8733 | -0.198 | -0.0923 | No |
| 82 | Chka | 8780 | -0.203 | -0.0902 | No |
| 83 | Sod2 | 8812 | -0.205 | -0.0867 | No |
| 84 | Rfc4 | 8971 | -0.216 | -0.0933 | No |
| 85 | Ppp1r2 | 9135 | -0.230 | -0.0998 | No |
| 86 | Lyn | 9410 | -0.251 | -0.1148 | No |
| 87 | Clcn2 | 9454 | -0.255 | -0.1109 | No |
| 88 | Gpx3 | 9580 | -0.267 | -0.1133 | No |
| 89 | Tfrc | 9637 | -0.270 | -0.1099 | No |
| 90 | Cdc5l | 9864 | -0.292 | -0.1198 | No |
| 91 | Selenow | 10007 | -0.301 | -0.1226 | No |
| 92 | Pole3 | 10252 | -0.322 | -0.1331 | No |
| 93 | E2f5 | 10444 | -0.341 | -0.1387 | No |
| 94 | H2ax | 10505 | -0.348 | -0.1334 | No |
| 95 | Urod | 10752 | -0.379 | -0.1424 | No |
| 96 | Bsg | 11162 | -0.438 | -0.1629 | No |
| 97 | Sigmar1 | 11177 | -0.440 | -0.1512 | No |
| 98 | Cltb | 11324 | -0.461 | -0.1496 | No |
| 99 | Tmbim6 | 11343 | -0.465 | -0.1374 | No |
| 100 | Pdap1 | 11393 | -0.473 | -0.1276 | No |
| 101 | Fen1 | 11471 | -0.488 | -0.1196 | No |
| 102 | Grina | 11484 | -0.489 | -0.1062 | No |
| 103 | Nat2 | 11522 | -0.497 | -0.0947 | No |
| 104 | Gls | 11634 | -0.522 | -0.0885 | No |
| 105 | Dnaja1 | 11712 | -0.538 | -0.0790 | No |
| 106 | Ppat | 11724 | -0.544 | -0.0640 | No |
| 107 | Hnrnpu | 11733 | -0.547 | -0.0486 | No |
| 108 | Ppif | 11919 | -0.609 | -0.0459 | No |
| 109 | Wiz | 11978 | -0.632 | -0.0322 | No |
| 110 | Tap1 | 12036 | -0.662 | -0.0174 | No |
| 111 | Bid | 12175 | -0.735 | -0.0072 | No |
| 112 | Furin | 12216 | -0.767 | 0.0120 | No |