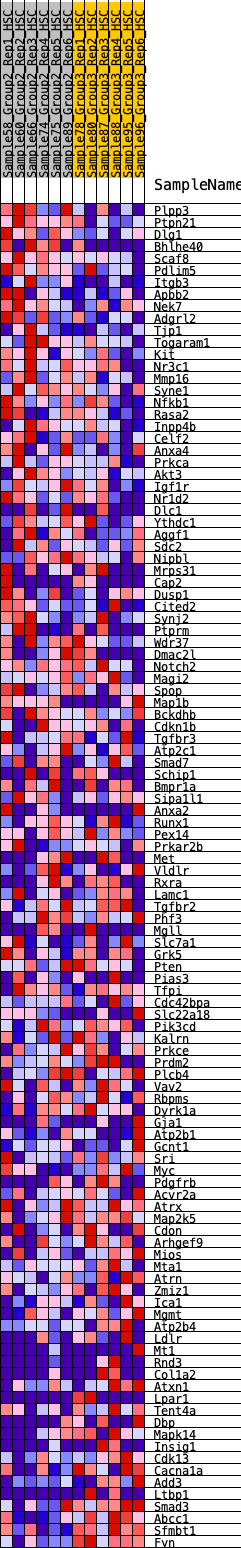
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
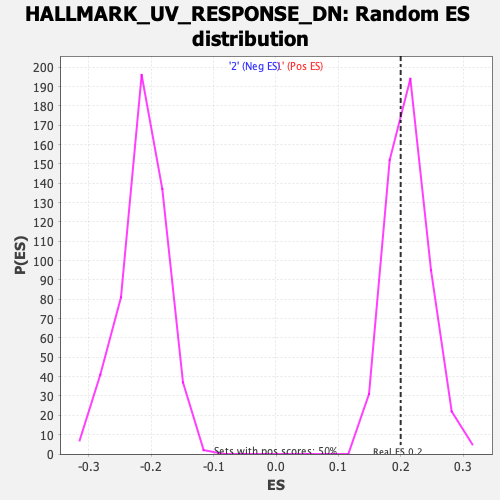
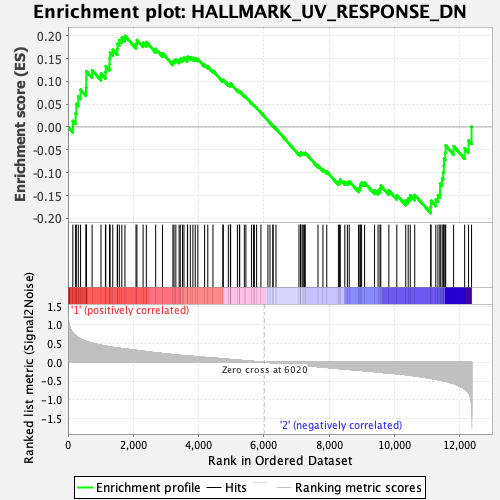
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.19954821 |
| Normalized Enrichment Score (NES) | 0.9463775 |
| Nominal p-value | 0.61122245 |
| FDR q-value | 0.6935956 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plpp3 | 148 | 0.785 | 0.0134 | Yes |
| 2 | Ptpn21 | 232 | 0.710 | 0.0296 | Yes |
| 3 | Dlg1 | 254 | 0.698 | 0.0505 | Yes |
| 4 | Bhlhe40 | 315 | 0.658 | 0.0670 | Yes |
| 5 | Scaf8 | 384 | 0.625 | 0.0817 | Yes |
| 6 | Pdlim5 | 551 | 0.563 | 0.0864 | Yes |
| 7 | Itgb3 | 560 | 0.561 | 0.1039 | Yes |
| 8 | Apbb2 | 563 | 0.560 | 0.1219 | Yes |
| 9 | Nek7 | 738 | 0.511 | 0.1243 | Yes |
| 10 | Adgrl2 | 1010 | 0.457 | 0.1170 | Yes |
| 11 | Tjp1 | 1146 | 0.432 | 0.1200 | Yes |
| 12 | Togaram1 | 1157 | 0.430 | 0.1331 | Yes |
| 13 | Kit | 1270 | 0.414 | 0.1374 | Yes |
| 14 | Nr3c1 | 1273 | 0.413 | 0.1507 | Yes |
| 15 | Mmp16 | 1289 | 0.410 | 0.1627 | Yes |
| 16 | Syne1 | 1370 | 0.399 | 0.1691 | Yes |
| 17 | Nfkb1 | 1508 | 0.383 | 0.1704 | Yes |
| 18 | Rasa2 | 1516 | 0.383 | 0.1822 | Yes |
| 19 | Inpp4b | 1571 | 0.376 | 0.1900 | Yes |
| 20 | Celf2 | 1648 | 0.366 | 0.1957 | Yes |
| 21 | Anxa4 | 1744 | 0.359 | 0.1995 | Yes |
| 22 | Prkca | 2082 | 0.321 | 0.1825 | No |
| 23 | Akt3 | 2110 | 0.317 | 0.1905 | No |
| 24 | Igf1r | 2299 | 0.297 | 0.1848 | No |
| 25 | Nr1d2 | 2398 | 0.285 | 0.1860 | No |
| 26 | Dlc1 | 2684 | 0.254 | 0.1710 | No |
| 27 | Ythdc1 | 2892 | 0.235 | 0.1617 | No |
| 28 | Aggf1 | 3212 | 0.205 | 0.1424 | No |
| 29 | Sdc2 | 3246 | 0.201 | 0.1462 | No |
| 30 | Nipbl | 3302 | 0.196 | 0.1480 | No |
| 31 | Mrps31 | 3402 | 0.189 | 0.1461 | No |
| 32 | Cap2 | 3437 | 0.185 | 0.1493 | No |
| 33 | Dusp1 | 3506 | 0.179 | 0.1496 | No |
| 34 | Cited2 | 3550 | 0.177 | 0.1518 | No |
| 35 | Synj2 | 3656 | 0.172 | 0.1488 | No |
| 36 | Ptprm | 3661 | 0.172 | 0.1540 | No |
| 37 | Wdr37 | 3744 | 0.164 | 0.1526 | No |
| 38 | Dmac2l | 3824 | 0.158 | 0.1513 | No |
| 39 | Notch2 | 3895 | 0.152 | 0.1505 | No |
| 40 | Magi2 | 3976 | 0.147 | 0.1488 | No |
| 41 | Spop | 4183 | 0.130 | 0.1362 | No |
| 42 | Map1b | 4280 | 0.123 | 0.1323 | No |
| 43 | Bckdhb | 4438 | 0.111 | 0.1231 | No |
| 44 | Cdkn1b | 4739 | 0.092 | 0.1016 | No |
| 45 | Tgfbr3 | 4755 | 0.090 | 0.1033 | No |
| 46 | Atp2c1 | 4908 | 0.079 | 0.0934 | No |
| 47 | Smad7 | 4971 | 0.075 | 0.0908 | No |
| 48 | Schip1 | 4975 | 0.075 | 0.0930 | No |
| 49 | Bmpr1a | 4979 | 0.074 | 0.0951 | No |
| 50 | Sipa1l1 | 5186 | 0.059 | 0.0802 | No |
| 51 | Anxa2 | 5248 | 0.054 | 0.0770 | No |
| 52 | Runx1 | 5258 | 0.054 | 0.0780 | No |
| 53 | Pex14 | 5399 | 0.043 | 0.0680 | No |
| 54 | Prkar2b | 5447 | 0.039 | 0.0654 | No |
| 55 | Met | 5620 | 0.027 | 0.0522 | No |
| 56 | Vldlr | 5681 | 0.023 | 0.0481 | No |
| 57 | Rxra | 5705 | 0.021 | 0.0469 | No |
| 58 | Lamc1 | 5775 | 0.017 | 0.0418 | No |
| 59 | Tgfbr2 | 5906 | 0.009 | 0.0315 | No |
| 60 | Phf3 | 6121 | -0.003 | 0.0141 | No |
| 61 | Mgll | 6184 | -0.007 | 0.0093 | No |
| 62 | Slc7a1 | 6272 | -0.013 | 0.0026 | No |
| 63 | Grk5 | 6278 | -0.013 | 0.0026 | No |
| 64 | Pten | 6368 | -0.019 | -0.0040 | No |
| 65 | Pias3 | 7071 | -0.069 | -0.0591 | No |
| 66 | Tfpi | 7118 | -0.073 | -0.0604 | No |
| 67 | Cdc42bpa | 7122 | -0.074 | -0.0583 | No |
| 68 | Slc22a18 | 7123 | -0.074 | -0.0559 | No |
| 69 | Pik3cd | 7170 | -0.077 | -0.0572 | No |
| 70 | Kalrn | 7203 | -0.079 | -0.0572 | No |
| 71 | Prkce | 7243 | -0.083 | -0.0577 | No |
| 72 | Prdm2 | 7264 | -0.085 | -0.0566 | No |
| 73 | Plcb4 | 7654 | -0.117 | -0.0846 | No |
| 74 | Vav2 | 7805 | -0.128 | -0.0927 | No |
| 75 | Rbpms | 7922 | -0.137 | -0.0977 | No |
| 76 | Dyrk1a | 8278 | -0.166 | -0.1213 | No |
| 77 | Gja1 | 8315 | -0.168 | -0.1188 | No |
| 78 | Atp2b1 | 8336 | -0.170 | -0.1149 | No |
| 79 | Gcnt1 | 8475 | -0.181 | -0.1203 | No |
| 80 | Sri | 8553 | -0.186 | -0.1206 | No |
| 81 | Myc | 8610 | -0.191 | -0.1190 | No |
| 82 | Pdgfrb | 8896 | -0.210 | -0.1354 | No |
| 83 | Acvr2a | 8938 | -0.213 | -0.1318 | No |
| 84 | Atrx | 8950 | -0.214 | -0.1258 | No |
| 85 | Map2k5 | 8989 | -0.217 | -0.1219 | No |
| 86 | Cdon | 9080 | -0.226 | -0.1219 | No |
| 87 | Arhgef9 | 9385 | -0.249 | -0.1387 | No |
| 88 | Mios | 9493 | -0.259 | -0.1390 | No |
| 89 | Mta1 | 9552 | -0.264 | -0.1352 | No |
| 90 | Atrn | 9577 | -0.267 | -0.1285 | No |
| 91 | Zmiz1 | 9823 | -0.287 | -0.1392 | No |
| 92 | Ica1 | 10067 | -0.306 | -0.1491 | No |
| 93 | Mgmt | 10340 | -0.330 | -0.1605 | No |
| 94 | Atp2b4 | 10415 | -0.339 | -0.1556 | No |
| 95 | Ldlr | 10478 | -0.346 | -0.1494 | No |
| 96 | Mt1 | 10615 | -0.361 | -0.1488 | No |
| 97 | Rnd3 | 11104 | -0.429 | -0.1748 | No |
| 98 | Col1a2 | 11114 | -0.430 | -0.1616 | No |
| 99 | Atxn1 | 11260 | -0.452 | -0.1587 | No |
| 100 | Lpar1 | 11326 | -0.462 | -0.1491 | No |
| 101 | Tent4a | 11390 | -0.473 | -0.1389 | No |
| 102 | Dbp | 11392 | -0.473 | -0.1236 | No |
| 103 | Mapk14 | 11453 | -0.485 | -0.1128 | No |
| 104 | Insig1 | 11488 | -0.490 | -0.0997 | No |
| 105 | Cdk13 | 11505 | -0.494 | -0.0850 | No |
| 106 | Cacna1a | 11510 | -0.495 | -0.0693 | No |
| 107 | Add3 | 11550 | -0.504 | -0.0561 | No |
| 108 | Ltbp1 | 11564 | -0.506 | -0.0408 | No |
| 109 | Smad3 | 11806 | -0.566 | -0.0421 | No |
| 110 | Abcc1 | 12144 | -0.714 | -0.0464 | No |
| 111 | Sfmbt1 | 12263 | -0.811 | -0.0298 | No |
| 112 | Fyn | 12355 | -1.167 | 0.0007 | No |