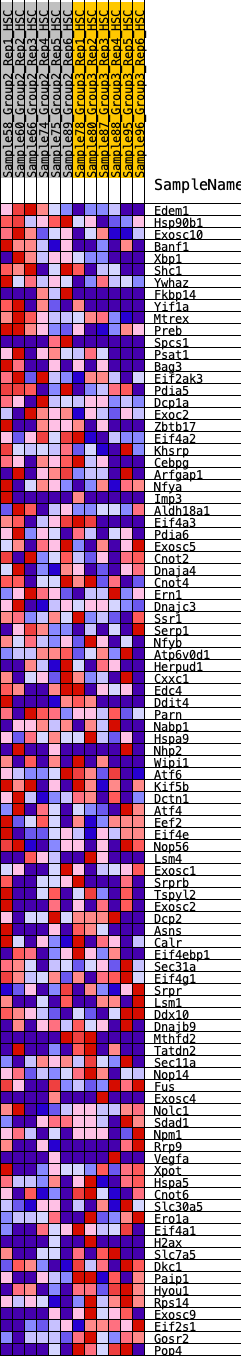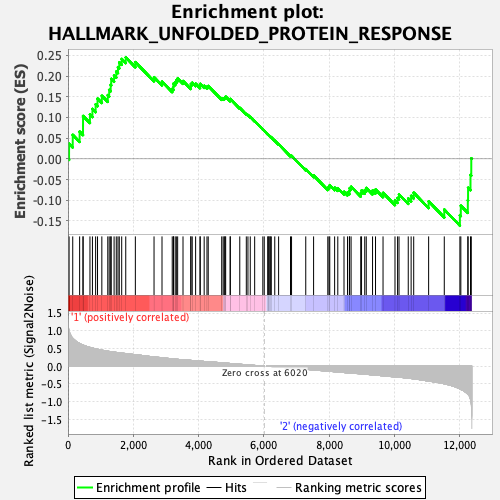
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.24521263 |
| Normalized Enrichment Score (NES) | 0.94847983 |
| Nominal p-value | 0.486 |
| FDR q-value | 0.72689575 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Edem1 | 33 | 1.011 | 0.0365 | Yes |
| 2 | Hsp90b1 | 145 | 0.786 | 0.0580 | Yes |
| 3 | Exosc10 | 356 | 0.642 | 0.0658 | Yes |
| 4 | Banf1 | 460 | 0.597 | 0.0805 | Yes |
| 5 | Xbp1 | 461 | 0.596 | 0.1037 | Yes |
| 6 | Shc1 | 672 | 0.529 | 0.1071 | Yes |
| 7 | Ywhaz | 748 | 0.509 | 0.1207 | Yes |
| 8 | Fkbp14 | 846 | 0.488 | 0.1317 | Yes |
| 9 | Yif1a | 904 | 0.478 | 0.1456 | Yes |
| 10 | Mtrex | 1036 | 0.452 | 0.1525 | Yes |
| 11 | Preb | 1218 | 0.422 | 0.1541 | Yes |
| 12 | Spcs1 | 1261 | 0.415 | 0.1668 | Yes |
| 13 | Psat1 | 1307 | 0.407 | 0.1789 | Yes |
| 14 | Bag3 | 1323 | 0.405 | 0.1934 | Yes |
| 15 | Eif2ak3 | 1411 | 0.393 | 0.2016 | Yes |
| 16 | Pdia5 | 1478 | 0.388 | 0.2112 | Yes |
| 17 | Dcp1a | 1530 | 0.380 | 0.2218 | Yes |
| 18 | Exoc2 | 1570 | 0.376 | 0.2332 | Yes |
| 19 | Zbtb17 | 1644 | 0.366 | 0.2415 | Yes |
| 20 | Eif4a2 | 1768 | 0.355 | 0.2452 | Yes |
| 21 | Khsrp | 2062 | 0.323 | 0.2339 | No |
| 22 | Cebpg | 2634 | 0.259 | 0.1974 | No |
| 23 | Arfgap1 | 2878 | 0.237 | 0.1867 | No |
| 24 | Nfya | 3191 | 0.207 | 0.1693 | No |
| 25 | Imp3 | 3228 | 0.203 | 0.1743 | No |
| 26 | Aldh18a1 | 3230 | 0.203 | 0.1820 | No |
| 27 | Eif4a3 | 3281 | 0.197 | 0.1856 | No |
| 28 | Pdia6 | 3317 | 0.194 | 0.1903 | No |
| 29 | Exosc5 | 3357 | 0.192 | 0.1946 | No |
| 30 | Cnot2 | 3521 | 0.178 | 0.1882 | No |
| 31 | Dnaja4 | 3760 | 0.162 | 0.1751 | No |
| 32 | Cnot4 | 3763 | 0.162 | 0.1812 | No |
| 33 | Ern1 | 3803 | 0.159 | 0.1842 | No |
| 34 | Dnajc3 | 3907 | 0.151 | 0.1817 | No |
| 35 | Ssr1 | 4042 | 0.142 | 0.1762 | No |
| 36 | Serp1 | 4048 | 0.141 | 0.1813 | No |
| 37 | Nfyb | 4169 | 0.131 | 0.1766 | No |
| 38 | Atp6v0d1 | 4255 | 0.124 | 0.1745 | No |
| 39 | Herpud1 | 4293 | 0.122 | 0.1762 | No |
| 40 | Cxxc1 | 4706 | 0.095 | 0.1463 | No |
| 41 | Edc4 | 4750 | 0.091 | 0.1463 | No |
| 42 | Ddit4 | 4785 | 0.088 | 0.1470 | No |
| 43 | Parn | 4807 | 0.086 | 0.1486 | No |
| 44 | Nabp1 | 4827 | 0.085 | 0.1504 | No |
| 45 | Hspa9 | 4966 | 0.075 | 0.1420 | No |
| 46 | Nhp2 | 4968 | 0.075 | 0.1448 | No |
| 47 | Wipi1 | 5259 | 0.054 | 0.1233 | No |
| 48 | Atf6 | 5459 | 0.038 | 0.1085 | No |
| 49 | Kif5b | 5509 | 0.035 | 0.1059 | No |
| 50 | Dctn1 | 5580 | 0.030 | 0.1013 | No |
| 51 | Atf4 | 5715 | 0.020 | 0.0912 | No |
| 52 | Eef2 | 5965 | 0.004 | 0.0711 | No |
| 53 | Eif4e | 6008 | 0.001 | 0.0677 | No |
| 54 | Nop56 | 6116 | -0.002 | 0.0590 | No |
| 55 | Lsm4 | 6129 | -0.003 | 0.0582 | No |
| 56 | Exosc1 | 6151 | -0.005 | 0.0567 | No |
| 57 | Srprb | 6182 | -0.007 | 0.0545 | No |
| 58 | Tspyl2 | 6193 | -0.007 | 0.0540 | No |
| 59 | Exosc2 | 6224 | -0.010 | 0.0519 | No |
| 60 | Dcp2 | 6330 | -0.016 | 0.0440 | No |
| 61 | Asns | 6448 | -0.025 | 0.0354 | No |
| 62 | Calr | 6812 | -0.051 | 0.0078 | No |
| 63 | Eif4ebp1 | 6844 | -0.053 | 0.0074 | No |
| 64 | Sec31a | 7279 | -0.086 | -0.0247 | No |
| 65 | Eif4g1 | 7519 | -0.106 | -0.0401 | No |
| 66 | Srpr | 7954 | -0.140 | -0.0700 | No |
| 67 | Lsm1 | 7991 | -0.142 | -0.0674 | No |
| 68 | Ddx10 | 8014 | -0.144 | -0.0636 | No |
| 69 | Dnajb9 | 8163 | -0.156 | -0.0696 | No |
| 70 | Mthfd2 | 8260 | -0.165 | -0.0711 | No |
| 71 | Tatdn2 | 8450 | -0.178 | -0.0796 | No |
| 72 | Sec11a | 8558 | -0.186 | -0.0810 | No |
| 73 | Nop14 | 8614 | -0.192 | -0.0781 | No |
| 74 | Fus | 8621 | -0.192 | -0.0711 | No |
| 75 | Exosc4 | 8667 | -0.194 | -0.0673 | No |
| 76 | Nolc1 | 8962 | -0.215 | -0.0829 | No |
| 77 | Sdad1 | 8987 | -0.217 | -0.0764 | No |
| 78 | Npm1 | 9085 | -0.226 | -0.0756 | No |
| 79 | Rrp9 | 9131 | -0.230 | -0.0703 | No |
| 80 | Vegfa | 9324 | -0.245 | -0.0765 | No |
| 81 | Xpot | 9415 | -0.252 | -0.0741 | No |
| 82 | Hspa5 | 9645 | -0.272 | -0.0822 | No |
| 83 | Cnot6 | 10011 | -0.301 | -0.1002 | No |
| 84 | Slc30a5 | 10090 | -0.309 | -0.0946 | No |
| 85 | Ero1a | 10134 | -0.313 | -0.0860 | No |
| 86 | Eif4a1 | 10417 | -0.339 | -0.0958 | No |
| 87 | H2ax | 10505 | -0.348 | -0.0894 | No |
| 88 | Slc7a5 | 10584 | -0.358 | -0.0819 | No |
| 89 | Dkc1 | 11041 | -0.420 | -0.1027 | No |
| 90 | Paip1 | 11520 | -0.496 | -0.1225 | No |
| 91 | Hyou1 | 11998 | -0.643 | -0.1364 | No |
| 92 | Rps14 | 12027 | -0.656 | -0.1133 | No |
| 93 | Exosc9 | 12241 | -0.788 | -0.1000 | No |
| 94 | Eif2s1 | 12249 | -0.794 | -0.0698 | No |
| 95 | Gosr2 | 12323 | -0.951 | -0.0388 | No |
| 96 | Pop4 | 12347 | -1.083 | 0.0013 | No |