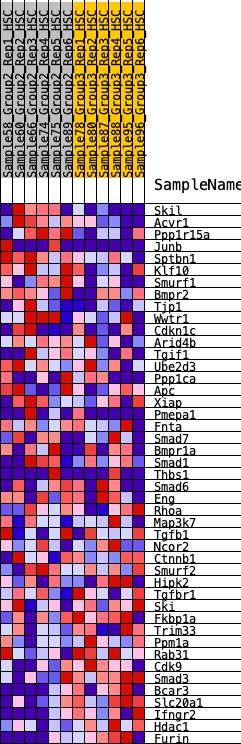
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
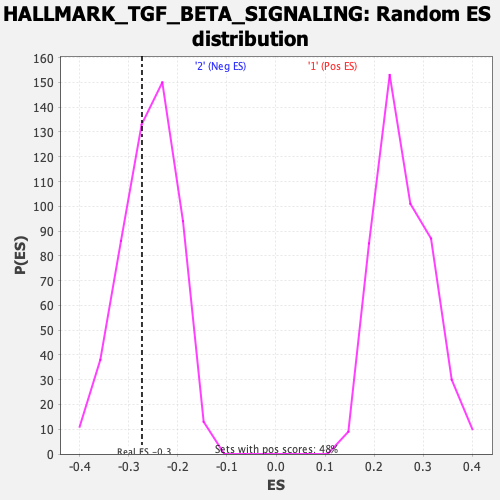
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.27290317 |
| Normalized Enrichment Score (NES) | -1.0547523 |
| Nominal p-value | 0.36190477 |
| FDR q-value | 0.61209375 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Skil | 173 | 0.762 | 0.0390 | Yes |
| 2 | Acvr1 | 195 | 0.742 | 0.0889 | Yes |
| 3 | Ppp1r15a | 425 | 0.607 | 0.1126 | Yes |
| 4 | Junb | 513 | 0.576 | 0.1456 | Yes |
| 5 | Sptbn1 | 847 | 0.488 | 0.1526 | Yes |
| 6 | Klf10 | 961 | 0.467 | 0.1759 | Yes |
| 7 | Smurf1 | 985 | 0.462 | 0.2061 | Yes |
| 8 | Bmpr2 | 988 | 0.462 | 0.2381 | Yes |
| 9 | Tjp1 | 1146 | 0.432 | 0.2555 | Yes |
| 10 | Wwtr1 | 1957 | 0.332 | 0.2128 | No |
| 11 | Cdkn1c | 2612 | 0.261 | 0.1779 | No |
| 12 | Arid4b | 2647 | 0.257 | 0.1931 | No |
| 13 | Tgif1 | 2710 | 0.252 | 0.2056 | No |
| 14 | Ube2d3 | 3122 | 0.214 | 0.1872 | No |
| 15 | Ppp1ca | 3277 | 0.198 | 0.1884 | No |
| 16 | Apc | 3526 | 0.178 | 0.1807 | No |
| 17 | Xiap | 3752 | 0.163 | 0.1737 | No |
| 18 | Pmepa1 | 3807 | 0.159 | 0.1804 | No |
| 19 | Fnta | 4671 | 0.098 | 0.1171 | No |
| 20 | Smad7 | 4971 | 0.075 | 0.0980 | No |
| 21 | Bmpr1a | 4979 | 0.074 | 0.1026 | No |
| 22 | Smad1 | 5135 | 0.061 | 0.0943 | No |
| 23 | Thbs1 | 5363 | 0.045 | 0.0791 | No |
| 24 | Smad6 | 5696 | 0.022 | 0.0536 | No |
| 25 | Eng | 5857 | 0.011 | 0.0415 | No |
| 26 | Rhoa | 5920 | 0.008 | 0.0369 | No |
| 27 | Map3k7 | 6195 | -0.007 | 0.0152 | No |
| 28 | Tgfb1 | 6512 | -0.030 | -0.0083 | No |
| 29 | Ncor2 | 7107 | -0.073 | -0.0515 | No |
| 30 | Ctnnb1 | 7135 | -0.074 | -0.0485 | No |
| 31 | Smurf2 | 7187 | -0.078 | -0.0472 | No |
| 32 | Hipk2 | 8339 | -0.170 | -0.1289 | No |
| 33 | Tgfbr1 | 8367 | -0.173 | -0.1190 | No |
| 34 | Ski | 9999 | -0.300 | -0.2305 | No |
| 35 | Fkbp1a | 10522 | -0.350 | -0.2485 | No |
| 36 | Trim33 | 10766 | -0.381 | -0.2417 | No |
| 37 | Ppm1a | 10932 | -0.404 | -0.2270 | No |
| 38 | Rab31 | 10995 | -0.415 | -0.2032 | No |
| 39 | Cdk9 | 11134 | -0.432 | -0.1843 | No |
| 40 | Smad3 | 11806 | -0.566 | -0.1994 | No |
| 41 | Bcar3 | 11848 | -0.581 | -0.1623 | No |
| 42 | Slc20a1 | 12117 | -0.698 | -0.1354 | No |
| 43 | Ifngr2 | 12120 | -0.699 | -0.0870 | No |
| 44 | Hdac1 | 12214 | -0.765 | -0.0413 | No |
| 45 | Furin | 12216 | -0.767 | 0.0119 | No |