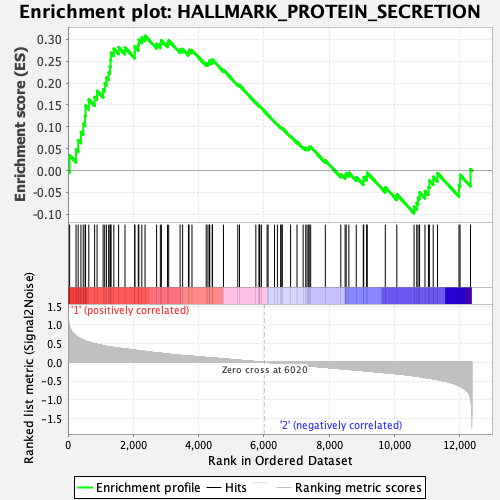
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.30805257 |
| Normalized Enrichment Score (NES) | 1.3229865 |
| Nominal p-value | 0.08565737 |
| FDR q-value | 0.3759685 |
| FWER p-Value | 0.715 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 50 | 0.947 | 0.0349 | Yes |
| 2 | Atp6v1h | 246 | 0.702 | 0.0478 | Yes |
| 3 | Sgms1 | 311 | 0.659 | 0.0697 | Yes |
| 4 | Ap3b1 | 396 | 0.620 | 0.0883 | Yes |
| 5 | Zw10 | 467 | 0.594 | 0.1070 | Yes |
| 6 | Snap23 | 523 | 0.573 | 0.1261 | Yes |
| 7 | Rps6ka3 | 537 | 0.568 | 0.1484 | Yes |
| 8 | Cln5 | 636 | 0.536 | 0.1625 | Yes |
| 9 | Yipf6 | 815 | 0.496 | 0.1683 | Yes |
| 10 | Mon2 | 890 | 0.481 | 0.1821 | Yes |
| 11 | Dst | 1075 | 0.445 | 0.1854 | Yes |
| 12 | Vps4b | 1124 | 0.436 | 0.1994 | Yes |
| 13 | Tom1l1 | 1176 | 0.426 | 0.2128 | Yes |
| 14 | Copb2 | 1245 | 0.419 | 0.2244 | Yes |
| 15 | Copb1 | 1283 | 0.412 | 0.2383 | Yes |
| 16 | Sec22b | 1305 | 0.408 | 0.2534 | Yes |
| 17 | Rab5a | 1317 | 0.406 | 0.2692 | Yes |
| 18 | Napg | 1403 | 0.394 | 0.2784 | Yes |
| 19 | Stx16 | 1550 | 0.378 | 0.2821 | Yes |
| 20 | Kif1b | 1745 | 0.359 | 0.2810 | Yes |
| 21 | Arf1 | 2044 | 0.325 | 0.2701 | Yes |
| 22 | Napa | 2045 | 0.325 | 0.2834 | Yes |
| 23 | Snx2 | 2152 | 0.312 | 0.2876 | Yes |
| 24 | Sspn | 2169 | 0.310 | 0.2991 | Yes |
| 25 | Vps45 | 2260 | 0.302 | 0.3041 | Yes |
| 26 | Mapk1 | 2359 | 0.290 | 0.3081 | Yes |
| 27 | Ykt6 | 2713 | 0.252 | 0.2897 | No |
| 28 | Clta | 2827 | 0.242 | 0.2904 | No |
| 29 | Arcn1 | 2859 | 0.238 | 0.2977 | No |
| 30 | Dop1a | 3045 | 0.220 | 0.2916 | No |
| 31 | Vamp4 | 3083 | 0.217 | 0.2975 | No |
| 32 | Rab14 | 3432 | 0.186 | 0.2768 | No |
| 33 | Arfip1 | 3509 | 0.179 | 0.2780 | No |
| 34 | Rab2a | 3687 | 0.168 | 0.2705 | No |
| 35 | Bet1 | 3705 | 0.167 | 0.2760 | No |
| 36 | Cope | 3796 | 0.160 | 0.2752 | No |
| 37 | Tmx1 | 4232 | 0.127 | 0.2450 | No |
| 38 | Krt18 | 4285 | 0.122 | 0.2457 | No |
| 39 | Adam10 | 4330 | 0.119 | 0.2470 | No |
| 40 | Cav2 | 4339 | 0.118 | 0.2512 | No |
| 41 | Rab9 | 4416 | 0.112 | 0.2496 | No |
| 42 | Ocrl | 4422 | 0.112 | 0.2538 | No |
| 43 | Uso1 | 4759 | 0.090 | 0.2302 | No |
| 44 | Scamp3 | 5198 | 0.058 | 0.1969 | No |
| 45 | Golga4 | 5250 | 0.054 | 0.1950 | No |
| 46 | Stx7 | 5751 | 0.018 | 0.1550 | No |
| 47 | M6pr | 5846 | 0.012 | 0.1478 | No |
| 48 | Stam | 5861 | 0.011 | 0.1471 | No |
| 49 | Cog2 | 5872 | 0.011 | 0.1468 | No |
| 50 | Ppt1 | 5921 | 0.008 | 0.1432 | No |
| 51 | Ergic3 | 6096 | -0.001 | 0.1290 | No |
| 52 | Ap1g1 | 6126 | -0.003 | 0.1268 | No |
| 53 | Stx12 | 6323 | -0.016 | 0.1115 | No |
| 54 | Bnip3 | 6411 | -0.022 | 0.1053 | No |
| 55 | Atp7a | 6511 | -0.030 | 0.0985 | No |
| 56 | Clcn3 | 6527 | -0.030 | 0.0985 | No |
| 57 | Lamp2 | 6563 | -0.034 | 0.0971 | No |
| 58 | Dnm1l | 6815 | -0.051 | 0.0787 | No |
| 59 | Scamp1 | 7011 | -0.066 | 0.0655 | No |
| 60 | Arfgef2 | 7199 | -0.079 | 0.0535 | No |
| 61 | Sec31a | 7279 | -0.086 | 0.0506 | No |
| 62 | Anp32e | 7320 | -0.090 | 0.0511 | No |
| 63 | Tmed10 | 7371 | -0.095 | 0.0509 | No |
| 64 | Tsg101 | 7382 | -0.096 | 0.0540 | No |
| 65 | Atp1a1 | 7430 | -0.099 | 0.0543 | No |
| 66 | Vamp3 | 7878 | -0.134 | 0.0234 | No |
| 67 | Sec24d | 8352 | -0.171 | -0.0081 | No |
| 68 | Ap2s1 | 8484 | -0.182 | -0.0113 | No |
| 69 | Ap2b1 | 8516 | -0.184 | -0.0063 | No |
| 70 | Ap3s1 | 8599 | -0.190 | -0.0052 | No |
| 71 | Ctsc | 8825 | -0.206 | -0.0150 | No |
| 72 | Scrn1 | 9040 | -0.222 | -0.0233 | No |
| 73 | Rab22a | 9054 | -0.223 | -0.0152 | No |
| 74 | Galc | 9142 | -0.231 | -0.0128 | No |
| 75 | Gbf1 | 9162 | -0.232 | -0.0048 | No |
| 76 | Cltc | 9714 | -0.279 | -0.0383 | No |
| 77 | Ica1 | 10067 | -0.306 | -0.0544 | No |
| 78 | Arfgap3 | 10595 | -0.359 | -0.0825 | No |
| 79 | Gnas | 10677 | -0.368 | -0.0740 | No |
| 80 | Lman1 | 10717 | -0.372 | -0.0619 | No |
| 81 | Rer1 | 10764 | -0.381 | -0.0499 | No |
| 82 | Tpd52 | 10929 | -0.404 | -0.0467 | No |
| 83 | Igf2r | 11035 | -0.419 | -0.0380 | No |
| 84 | Pam | 11064 | -0.423 | -0.0229 | No |
| 85 | Ap2m1 | 11181 | -0.441 | -0.0143 | No |
| 86 | Arfgef1 | 11312 | -0.459 | -0.0060 | No |
| 87 | Cd63 | 11970 | -0.627 | -0.0337 | No |
| 88 | Sod1 | 12003 | -0.643 | -0.0099 | No |
| 89 | Gosr2 | 12323 | -0.951 | 0.0033 | No |