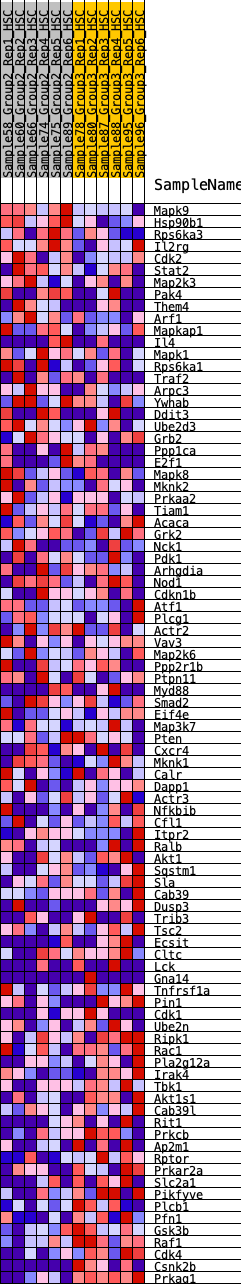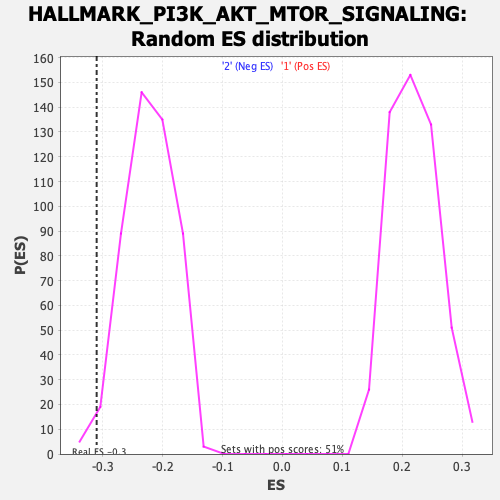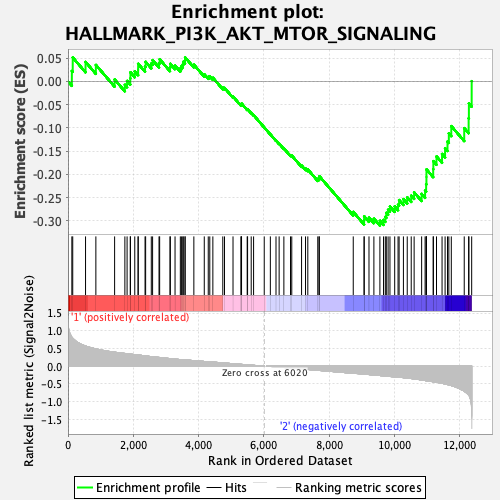
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.31003764 |
| Normalized Enrichment Score (NES) | -1.4030885 |
| Nominal p-value | 0.020576132 |
| FDR q-value | 0.86957216 |
| FWER p-Value | 0.537 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mapk9 | 118 | 0.815 | 0.0225 | No |
| 2 | Hsp90b1 | 145 | 0.786 | 0.0513 | No |
| 3 | Rps6ka3 | 537 | 0.568 | 0.0418 | No |
| 4 | Il2rg | 853 | 0.487 | 0.0353 | No |
| 5 | Cdk2 | 1427 | 0.392 | 0.0040 | No |
| 6 | Stat2 | 1741 | 0.359 | -0.0073 | No |
| 7 | Map2k3 | 1809 | 0.350 | 0.0010 | No |
| 8 | Pak4 | 1907 | 0.339 | 0.0064 | No |
| 9 | Them4 | 1911 | 0.338 | 0.0195 | No |
| 10 | Arf1 | 2044 | 0.325 | 0.0215 | No |
| 11 | Mapkap1 | 2147 | 0.312 | 0.0255 | No |
| 12 | Il4 | 2148 | 0.312 | 0.0377 | No |
| 13 | Mapk1 | 2359 | 0.290 | 0.0320 | No |
| 14 | Rps6ka1 | 2377 | 0.287 | 0.0420 | No |
| 15 | Traf2 | 2550 | 0.267 | 0.0385 | No |
| 16 | Arpc3 | 2590 | 0.263 | 0.0456 | No |
| 17 | Ywhab | 2787 | 0.247 | 0.0394 | No |
| 18 | Ddit3 | 2806 | 0.244 | 0.0475 | No |
| 19 | Ube2d3 | 3122 | 0.214 | 0.0303 | No |
| 20 | Grb2 | 3133 | 0.213 | 0.0378 | No |
| 21 | Ppp1ca | 3277 | 0.198 | 0.0340 | No |
| 22 | E2f1 | 3439 | 0.185 | 0.0281 | No |
| 23 | Mapk8 | 3472 | 0.183 | 0.0327 | No |
| 24 | Mknk2 | 3514 | 0.179 | 0.0364 | No |
| 25 | Prkaa2 | 3530 | 0.178 | 0.0422 | No |
| 26 | Tiam1 | 3582 | 0.174 | 0.0449 | No |
| 27 | Acaca | 3586 | 0.174 | 0.0515 | No |
| 28 | Grk2 | 3853 | 0.155 | 0.0359 | No |
| 29 | Nck1 | 4172 | 0.131 | 0.0151 | No |
| 30 | Pdk1 | 4294 | 0.121 | 0.0101 | No |
| 31 | Arhgdia | 4341 | 0.118 | 0.0109 | No |
| 32 | Nod1 | 4435 | 0.111 | 0.0077 | No |
| 33 | Cdkn1b | 4739 | 0.092 | -0.0133 | No |
| 34 | Atf1 | 4791 | 0.088 | -0.0140 | No |
| 35 | Plcg1 | 5052 | 0.068 | -0.0325 | No |
| 36 | Actr2 | 5295 | 0.051 | -0.0503 | No |
| 37 | Vav3 | 5306 | 0.050 | -0.0491 | No |
| 38 | Map2k6 | 5313 | 0.050 | -0.0476 | No |
| 39 | Ppp2r1b | 5486 | 0.036 | -0.0602 | No |
| 40 | Ptpn11 | 5498 | 0.035 | -0.0598 | No |
| 41 | Myd88 | 5609 | 0.028 | -0.0676 | No |
| 42 | Smad2 | 5682 | 0.023 | -0.0726 | No |
| 43 | Eif4e | 6008 | 0.001 | -0.0990 | No |
| 44 | Map3k7 | 6195 | -0.007 | -0.1139 | No |
| 45 | Pten | 6368 | -0.019 | -0.1272 | No |
| 46 | Cxcr4 | 6464 | -0.026 | -0.1339 | No |
| 47 | Mknk1 | 6609 | -0.037 | -0.1441 | No |
| 48 | Calr | 6812 | -0.051 | -0.1586 | No |
| 49 | Dapp1 | 6851 | -0.054 | -0.1596 | No |
| 50 | Actr3 | 7151 | -0.076 | -0.1809 | No |
| 51 | Nfkbib | 7272 | -0.086 | -0.1873 | No |
| 52 | Cfl1 | 7342 | -0.092 | -0.1893 | No |
| 53 | Itpr2 | 7647 | -0.116 | -0.2095 | No |
| 54 | Ralb | 7685 | -0.119 | -0.2079 | No |
| 55 | Akt1 | 7695 | -0.120 | -0.2039 | No |
| 56 | Sqstm1 | 8733 | -0.198 | -0.2806 | No |
| 57 | Sla | 9065 | -0.223 | -0.2988 | No |
| 58 | Cab39 | 9068 | -0.224 | -0.2901 | No |
| 59 | Dusp3 | 9215 | -0.236 | -0.2927 | No |
| 60 | Trib3 | 9365 | -0.247 | -0.2951 | No |
| 61 | Tsc2 | 9549 | -0.264 | -0.2997 | Yes |
| 62 | Ecsit | 9660 | -0.274 | -0.2979 | Yes |
| 63 | Cltc | 9714 | -0.279 | -0.2912 | Yes |
| 64 | Lck | 9745 | -0.281 | -0.2826 | Yes |
| 65 | Gna14 | 9800 | -0.286 | -0.2758 | Yes |
| 66 | Tnfrsf1a | 9857 | -0.291 | -0.2689 | Yes |
| 67 | Pin1 | 10002 | -0.300 | -0.2688 | Yes |
| 68 | Cdk1 | 10105 | -0.310 | -0.2649 | Yes |
| 69 | Ube2n | 10137 | -0.313 | -0.2551 | Yes |
| 70 | Ripk1 | 10269 | -0.324 | -0.2531 | Yes |
| 71 | Rac1 | 10386 | -0.333 | -0.2494 | Yes |
| 72 | Pla2g12a | 10508 | -0.348 | -0.2455 | Yes |
| 73 | Irak4 | 10597 | -0.359 | -0.2386 | Yes |
| 74 | Tbk1 | 10827 | -0.389 | -0.2419 | Yes |
| 75 | Akt1s1 | 10933 | -0.404 | -0.2346 | Yes |
| 76 | Cab39l | 10965 | -0.410 | -0.2210 | Yes |
| 77 | Rit1 | 10974 | -0.411 | -0.2054 | Yes |
| 78 | Prkcb | 10975 | -0.411 | -0.1892 | Yes |
| 79 | Ap2m1 | 11181 | -0.441 | -0.1886 | Yes |
| 80 | Rptor | 11186 | -0.442 | -0.1715 | Yes |
| 81 | Prkar2a | 11282 | -0.455 | -0.1613 | Yes |
| 82 | Slc2a1 | 11452 | -0.485 | -0.1560 | Yes |
| 83 | Pikfyve | 11545 | -0.502 | -0.1438 | Yes |
| 84 | Plcb1 | 11619 | -0.519 | -0.1293 | Yes |
| 85 | Pfn1 | 11660 | -0.529 | -0.1117 | Yes |
| 86 | Gsk3b | 11735 | -0.548 | -0.0962 | Yes |
| 87 | Raf1 | 12131 | -0.707 | -0.1006 | Yes |
| 88 | Cdk4 | 12266 | -0.814 | -0.0795 | Yes |
| 89 | Csnk2b | 12274 | -0.825 | -0.0476 | Yes |
| 90 | Prkag1 | 12359 | -1.392 | 0.0003 | Yes |