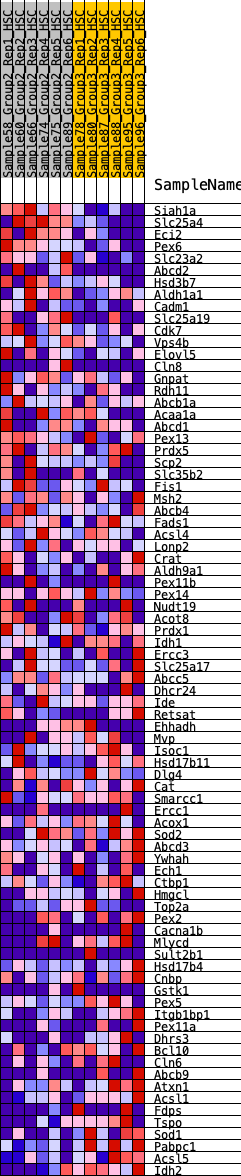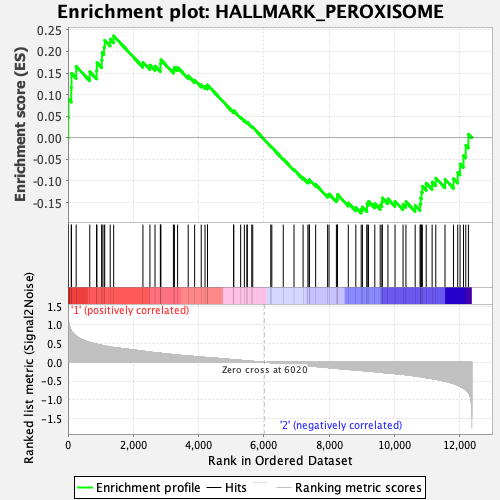
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.23601758 |
| Normalized Enrichment Score (NES) | 1.0093119 |
| Nominal p-value | 0.43912175 |
| FDR q-value | 0.64475924 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Siah1a | 6 | 1.179 | 0.0467 | Yes |
| 2 | Slc25a4 | 15 | 1.084 | 0.0894 | Yes |
| 3 | Eci2 | 99 | 0.841 | 0.1163 | Yes |
| 4 | Pex6 | 107 | 0.834 | 0.1491 | Yes |
| 5 | Slc23a2 | 250 | 0.700 | 0.1656 | Yes |
| 6 | Abcd2 | 666 | 0.530 | 0.1530 | Yes |
| 7 | Hsd3b7 | 874 | 0.484 | 0.1555 | Yes |
| 8 | Aldh1a1 | 885 | 0.482 | 0.1740 | Yes |
| 9 | Cadm1 | 1031 | 0.452 | 0.1803 | Yes |
| 10 | Slc25a19 | 1044 | 0.450 | 0.1973 | Yes |
| 11 | Cdk7 | 1103 | 0.439 | 0.2102 | Yes |
| 12 | Vps4b | 1124 | 0.436 | 0.2260 | Yes |
| 13 | Elovl5 | 1292 | 0.409 | 0.2288 | Yes |
| 14 | Cln8 | 1398 | 0.395 | 0.2360 | Yes |
| 15 | Gnpat | 2294 | 0.297 | 0.1750 | No |
| 16 | Rdh11 | 2508 | 0.272 | 0.1686 | No |
| 17 | Abcb1a | 2664 | 0.256 | 0.1662 | No |
| 18 | Acaa1a | 2829 | 0.242 | 0.1625 | No |
| 19 | Abcd1 | 2830 | 0.241 | 0.1722 | No |
| 20 | Pex13 | 2844 | 0.240 | 0.1807 | No |
| 21 | Prdx5 | 3227 | 0.203 | 0.1577 | No |
| 22 | Scp2 | 3258 | 0.200 | 0.1633 | No |
| 23 | Slc35b2 | 3355 | 0.192 | 0.1632 | No |
| 24 | Fis1 | 3680 | 0.169 | 0.1435 | No |
| 25 | Msh2 | 3877 | 0.153 | 0.1337 | No |
| 26 | Abcb4 | 4079 | 0.139 | 0.1229 | No |
| 27 | Fads1 | 4195 | 0.129 | 0.1187 | No |
| 28 | Acsl4 | 4270 | 0.123 | 0.1176 | No |
| 29 | Lonp2 | 4271 | 0.123 | 0.1225 | No |
| 30 | Crat | 5071 | 0.066 | 0.0602 | No |
| 31 | Aldh9a1 | 5074 | 0.066 | 0.0626 | No |
| 32 | Pex11b | 5286 | 0.051 | 0.0475 | No |
| 33 | Pex14 | 5399 | 0.043 | 0.0401 | No |
| 34 | Nudt19 | 5477 | 0.037 | 0.0353 | No |
| 35 | Acot8 | 5489 | 0.036 | 0.0358 | No |
| 36 | Prdx1 | 5626 | 0.027 | 0.0258 | No |
| 37 | Idh1 | 5656 | 0.025 | 0.0244 | No |
| 38 | Ercc3 | 6208 | -0.009 | -0.0201 | No |
| 39 | Slc25a17 | 6240 | -0.011 | -0.0222 | No |
| 40 | Abcc5 | 6590 | -0.036 | -0.0491 | No |
| 41 | Dhcr24 | 6919 | -0.059 | -0.0735 | No |
| 42 | Ide | 7197 | -0.079 | -0.0929 | No |
| 43 | Retsat | 7345 | -0.093 | -0.1012 | No |
| 44 | Ehhadh | 7385 | -0.096 | -0.1005 | No |
| 45 | Mvp | 7387 | -0.096 | -0.0967 | No |
| 46 | Isoc1 | 7582 | -0.110 | -0.1081 | No |
| 47 | Hsd17b11 | 7948 | -0.139 | -0.1323 | No |
| 48 | Dlg4 | 7992 | -0.142 | -0.1301 | No |
| 49 | Cat | 8217 | -0.160 | -0.1419 | No |
| 50 | Smarcc1 | 8245 | -0.163 | -0.1376 | No |
| 51 | Ercc1 | 8248 | -0.164 | -0.1312 | No |
| 52 | Acox1 | 8581 | -0.188 | -0.1507 | No |
| 53 | Sod2 | 8812 | -0.205 | -0.1612 | No |
| 54 | Abcd3 | 8976 | -0.216 | -0.1658 | No |
| 55 | Ywhah | 9014 | -0.219 | -0.1600 | No |
| 56 | Ech1 | 9153 | -0.232 | -0.1620 | No |
| 57 | Ctbp1 | 9154 | -0.232 | -0.1527 | No |
| 58 | Hmgcl | 9201 | -0.235 | -0.1471 | No |
| 59 | Top2a | 9393 | -0.250 | -0.1526 | No |
| 60 | Pex2 | 9558 | -0.265 | -0.1554 | No |
| 61 | Cacna1b | 9611 | -0.268 | -0.1489 | No |
| 62 | Mlycd | 9623 | -0.269 | -0.1390 | No |
| 63 | Sult2b1 | 9795 | -0.286 | -0.1415 | No |
| 64 | Hsd17b4 | 10012 | -0.302 | -0.1470 | No |
| 65 | Cnbp | 10258 | -0.323 | -0.1540 | No |
| 66 | Gstk1 | 10346 | -0.331 | -0.1479 | No |
| 67 | Pex5 | 10629 | -0.362 | -0.1563 | No |
| 68 | Itgb1bp1 | 10778 | -0.383 | -0.1531 | No |
| 69 | Pex11a | 10799 | -0.385 | -0.1393 | No |
| 70 | Dhrs3 | 10824 | -0.389 | -0.1257 | No |
| 71 | Bcl10 | 10848 | -0.392 | -0.1119 | No |
| 72 | Cln6 | 10967 | -0.411 | -0.1051 | No |
| 73 | Abcb9 | 11148 | -0.435 | -0.1023 | No |
| 74 | Atxn1 | 11260 | -0.452 | -0.0932 | No |
| 75 | Acsl1 | 11542 | -0.501 | -0.0961 | No |
| 76 | Fdps | 11803 | -0.565 | -0.0946 | No |
| 77 | Tspo | 11932 | -0.613 | -0.0805 | No |
| 78 | Sod1 | 12003 | -0.643 | -0.0605 | No |
| 79 | Pabpc1 | 12108 | -0.693 | -0.0412 | No |
| 80 | Acsl5 | 12178 | -0.737 | -0.0173 | No |
| 81 | Idh2 | 12259 | -0.807 | 0.0085 | No |