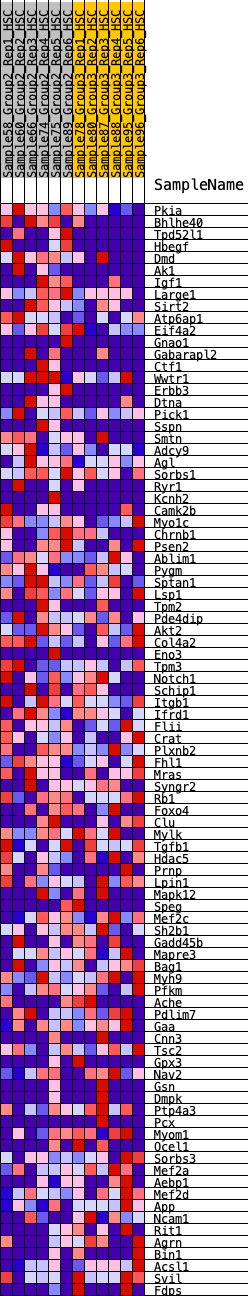
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
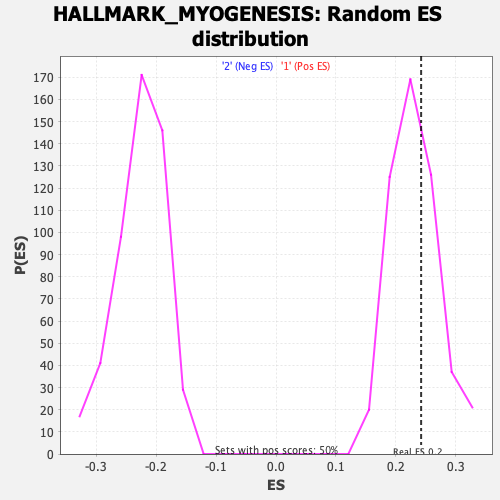
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.24206862 |
| Normalized Enrichment Score (NES) | 1.0559584 |
| Nominal p-value | 0.3493976 |
| FDR q-value | 0.5545279 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pkia | 266 | 0.692 | 0.0095 | Yes |
| 2 | Bhlhe40 | 315 | 0.658 | 0.0353 | Yes |
| 3 | Tpd52l1 | 438 | 0.604 | 0.0526 | Yes |
| 4 | Hbegf | 652 | 0.533 | 0.0593 | Yes |
| 5 | Dmd | 962 | 0.466 | 0.0552 | Yes |
| 6 | Ak1 | 977 | 0.464 | 0.0750 | Yes |
| 7 | Igf1 | 1113 | 0.438 | 0.0837 | Yes |
| 8 | Large1 | 1180 | 0.425 | 0.0975 | Yes |
| 9 | Sirt2 | 1276 | 0.413 | 0.1084 | Yes |
| 10 | Atp6ap1 | 1638 | 0.366 | 0.0955 | Yes |
| 11 | Eif4a2 | 1768 | 0.355 | 0.1010 | Yes |
| 12 | Gnao1 | 1786 | 0.354 | 0.1156 | Yes |
| 13 | Gabarapl2 | 1810 | 0.350 | 0.1295 | Yes |
| 14 | Ctf1 | 1819 | 0.349 | 0.1446 | Yes |
| 15 | Wwtr1 | 1957 | 0.332 | 0.1485 | Yes |
| 16 | Erbb3 | 1969 | 0.332 | 0.1626 | Yes |
| 17 | Dtna | 2011 | 0.329 | 0.1741 | Yes |
| 18 | Pick1 | 2155 | 0.311 | 0.1765 | Yes |
| 19 | Sspn | 2169 | 0.310 | 0.1894 | Yes |
| 20 | Smtn | 2191 | 0.310 | 0.2017 | Yes |
| 21 | Adcy9 | 2238 | 0.304 | 0.2117 | Yes |
| 22 | Agl | 2261 | 0.302 | 0.2235 | Yes |
| 23 | Sorbs1 | 2346 | 0.291 | 0.2298 | Yes |
| 24 | Ryr1 | 2357 | 0.290 | 0.2421 | Yes |
| 25 | Kcnh2 | 3108 | 0.215 | 0.1907 | No |
| 26 | Camk2b | 3203 | 0.206 | 0.1923 | No |
| 27 | Myo1c | 3222 | 0.204 | 0.2000 | No |
| 28 | Chrnb1 | 3416 | 0.188 | 0.1928 | No |
| 29 | Psen2 | 3751 | 0.163 | 0.1729 | No |
| 30 | Ablim1 | 3761 | 0.162 | 0.1795 | No |
| 31 | Pygm | 3777 | 0.161 | 0.1855 | No |
| 32 | Sptan1 | 3873 | 0.153 | 0.1847 | No |
| 33 | Lsp1 | 4052 | 0.141 | 0.1766 | No |
| 34 | Tpm2 | 4097 | 0.137 | 0.1792 | No |
| 35 | Pde4dip | 4160 | 0.133 | 0.1801 | No |
| 36 | Akt2 | 4236 | 0.126 | 0.1797 | No |
| 37 | Col4a2 | 4359 | 0.116 | 0.1750 | No |
| 38 | Eno3 | 4478 | 0.110 | 0.1704 | No |
| 39 | Tpm3 | 4530 | 0.108 | 0.1711 | No |
| 40 | Notch1 | 4886 | 0.080 | 0.1458 | No |
| 41 | Schip1 | 4975 | 0.075 | 0.1420 | No |
| 42 | Itgb1 | 5000 | 0.072 | 0.1433 | No |
| 43 | Ifrd1 | 5022 | 0.071 | 0.1448 | No |
| 44 | Flii | 5057 | 0.068 | 0.1450 | No |
| 45 | Crat | 5071 | 0.066 | 0.1470 | No |
| 46 | Plxnb2 | 5496 | 0.035 | 0.1140 | No |
| 47 | Fhl1 | 5605 | 0.028 | 0.1065 | No |
| 48 | Mras | 5647 | 0.025 | 0.1043 | No |
| 49 | Syngr2 | 6132 | -0.003 | 0.0650 | No |
| 50 | Rb1 | 6157 | -0.005 | 0.0632 | No |
| 51 | Foxo4 | 6194 | -0.007 | 0.0606 | No |
| 52 | Clu | 6467 | -0.026 | 0.0397 | No |
| 53 | Mylk | 6484 | -0.028 | 0.0396 | No |
| 54 | Tgfb1 | 6512 | -0.030 | 0.0388 | No |
| 55 | Hdac5 | 6525 | -0.030 | 0.0392 | No |
| 56 | Prnp | 6666 | -0.042 | 0.0296 | No |
| 57 | Lpin1 | 6792 | -0.050 | 0.0217 | No |
| 58 | Mapk12 | 7386 | -0.096 | -0.0223 | No |
| 59 | Speg | 7414 | -0.098 | -0.0200 | No |
| 60 | Mef2c | 7572 | -0.110 | -0.0279 | No |
| 61 | Sh2b1 | 7691 | -0.119 | -0.0321 | No |
| 62 | Gadd45b | 7747 | -0.124 | -0.0310 | No |
| 63 | Mapre3 | 7823 | -0.130 | -0.0312 | No |
| 64 | Bag1 | 7923 | -0.137 | -0.0331 | No |
| 65 | Myh9 | 8165 | -0.156 | -0.0457 | No |
| 66 | Pfkm | 8238 | -0.162 | -0.0443 | No |
| 67 | Ache | 8295 | -0.167 | -0.0413 | No |
| 68 | Pdlim7 | 8351 | -0.171 | -0.0381 | No |
| 69 | Gaa | 8606 | -0.191 | -0.0502 | No |
| 70 | Cnn3 | 8677 | -0.194 | -0.0471 | No |
| 71 | Tsc2 | 9549 | -0.264 | -0.1062 | No |
| 72 | Gpx3 | 9580 | -0.267 | -0.0966 | No |
| 73 | Nav2 | 9719 | -0.279 | -0.0952 | No |
| 74 | Gsn | 9782 | -0.285 | -0.0874 | No |
| 75 | Dmpk | 9784 | -0.285 | -0.0746 | No |
| 76 | Ptp4a3 | 9915 | -0.295 | -0.0719 | No |
| 77 | Pcx | 10103 | -0.310 | -0.0732 | No |
| 78 | Myom1 | 10204 | -0.318 | -0.0670 | No |
| 79 | Ocel1 | 10276 | -0.325 | -0.0581 | No |
| 80 | Sorbs3 | 10304 | -0.327 | -0.0455 | No |
| 81 | Mef2a | 10379 | -0.332 | -0.0366 | No |
| 82 | Aebp1 | 10490 | -0.347 | -0.0299 | No |
| 83 | Mef2d | 10608 | -0.361 | -0.0231 | No |
| 84 | App | 10680 | -0.368 | -0.0123 | No |
| 85 | Ncam1 | 10968 | -0.411 | -0.0172 | No |
| 86 | Rit1 | 10974 | -0.411 | 0.0010 | No |
| 87 | Agrn | 11060 | -0.423 | 0.0131 | No |
| 88 | Bin1 | 11127 | -0.432 | 0.0272 | No |
| 89 | Acsl1 | 11542 | -0.501 | 0.0161 | No |
| 90 | Svil | 11775 | -0.557 | 0.0223 | No |
| 91 | Fdps | 11803 | -0.565 | 0.0456 | No |