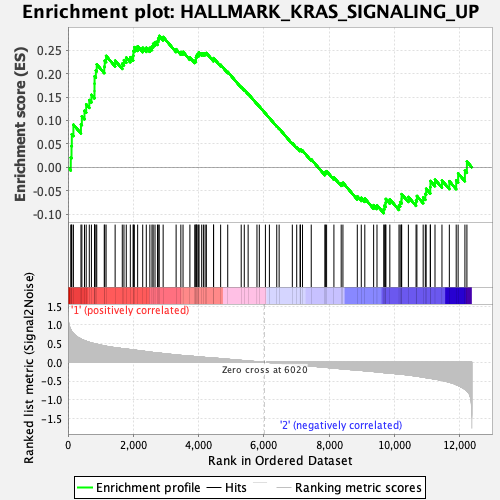
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.28064957 |
| Normalized Enrichment Score (NES) | 1.2494273 |
| Nominal p-value | 0.107942976 |
| FDR q-value | 0.37055647 |
| FWER p-Value | 0.861 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Birc3 | 85 | 0.873 | 0.0207 | Yes |
| 2 | Spon1 | 108 | 0.833 | 0.0453 | Yes |
| 3 | Dnmbp | 117 | 0.815 | 0.0705 | Yes |
| 4 | F13a1 | 166 | 0.769 | 0.0910 | Yes |
| 5 | Adam17 | 397 | 0.619 | 0.0918 | Yes |
| 6 | Ppp1r15a | 425 | 0.607 | 0.1089 | Yes |
| 7 | Satb1 | 505 | 0.578 | 0.1207 | Yes |
| 8 | Crot | 557 | 0.562 | 0.1344 | Yes |
| 9 | Hbegf | 652 | 0.533 | 0.1436 | Yes |
| 10 | St6gal1 | 717 | 0.515 | 0.1547 | Yes |
| 11 | Pdcd1lg2 | 811 | 0.496 | 0.1629 | Yes |
| 12 | Ets1 | 812 | 0.496 | 0.1786 | Yes |
| 13 | Kcnn4 | 814 | 0.496 | 0.1942 | Yes |
| 14 | Il2rg | 853 | 0.487 | 0.2066 | Yes |
| 15 | Gadd45g | 880 | 0.483 | 0.2197 | Yes |
| 16 | Il1rl2 | 1110 | 0.438 | 0.2149 | Yes |
| 17 | Trib1 | 1125 | 0.436 | 0.2276 | Yes |
| 18 | Klf4 | 1166 | 0.428 | 0.2379 | Yes |
| 19 | Vwa5a | 1443 | 0.391 | 0.2278 | Yes |
| 20 | Ephb2 | 1662 | 0.364 | 0.2215 | Yes |
| 21 | Epb41l3 | 1713 | 0.361 | 0.2289 | Yes |
| 22 | Ptbp2 | 1784 | 0.354 | 0.2344 | Yes |
| 23 | Plaur | 1908 | 0.339 | 0.2351 | Yes |
| 24 | Map4k1 | 1995 | 0.331 | 0.2385 | Yes |
| 25 | Gng11 | 2002 | 0.330 | 0.2485 | Yes |
| 26 | Cbr4 | 2031 | 0.326 | 0.2566 | Yes |
| 27 | Tmem176b | 2132 | 0.315 | 0.2584 | Yes |
| 28 | Evi5 | 2287 | 0.299 | 0.2553 | Yes |
| 29 | Strn | 2397 | 0.285 | 0.2554 | Yes |
| 30 | Tnfrsf1b | 2506 | 0.272 | 0.2552 | Yes |
| 31 | Laptm5 | 2571 | 0.266 | 0.2584 | Yes |
| 32 | Tspan13 | 2607 | 0.261 | 0.2639 | Yes |
| 33 | Abcb1a | 2664 | 0.256 | 0.2674 | Yes |
| 34 | Jup | 2740 | 0.250 | 0.2692 | Yes |
| 35 | Pecam1 | 2762 | 0.247 | 0.2753 | Yes |
| 36 | Adgrl4 | 2793 | 0.246 | 0.2806 | Yes |
| 37 | Tor1aip2 | 2913 | 0.233 | 0.2783 | No |
| 38 | Il10ra | 3310 | 0.194 | 0.2522 | No |
| 39 | Gucy1a1 | 3455 | 0.184 | 0.2463 | No |
| 40 | Mtmr10 | 3522 | 0.178 | 0.2465 | No |
| 41 | Tmem176a | 3732 | 0.165 | 0.2347 | No |
| 42 | Usp12 | 3881 | 0.153 | 0.2275 | No |
| 43 | Ammecr1 | 3917 | 0.151 | 0.2294 | No |
| 44 | Itgb2 | 3918 | 0.151 | 0.2342 | No |
| 45 | Inhba | 3930 | 0.150 | 0.2380 | No |
| 46 | Lcp1 | 3954 | 0.148 | 0.2408 | No |
| 47 | Mmd | 4004 | 0.144 | 0.2414 | No |
| 48 | Btc | 4007 | 0.144 | 0.2458 | No |
| 49 | Avl9 | 4093 | 0.137 | 0.2432 | No |
| 50 | Hdac9 | 4146 | 0.134 | 0.2432 | No |
| 51 | Rabgap1l | 4197 | 0.129 | 0.2432 | No |
| 52 | Akt2 | 4236 | 0.126 | 0.2441 | No |
| 53 | Ly96 | 4455 | 0.110 | 0.2298 | No |
| 54 | Kif5c | 4458 | 0.110 | 0.2332 | No |
| 55 | Atg10 | 4673 | 0.097 | 0.2188 | No |
| 56 | Rbm4 | 4890 | 0.080 | 0.2037 | No |
| 57 | Sdccag8 | 5303 | 0.050 | 0.1717 | No |
| 58 | Ccser2 | 5397 | 0.043 | 0.1655 | No |
| 59 | Mycn | 5515 | 0.034 | 0.1570 | No |
| 60 | Adgra2 | 5785 | 0.016 | 0.1356 | No |
| 61 | Eng | 5857 | 0.011 | 0.1302 | No |
| 62 | Tmem158 | 6044 | 0.000 | 0.1150 | No |
| 63 | Bpgm | 6166 | -0.006 | 0.1053 | No |
| 64 | Map7 | 6391 | -0.021 | 0.0877 | No |
| 65 | Cxcr4 | 6464 | -0.026 | 0.0826 | No |
| 66 | Cd37 | 6868 | -0.055 | 0.0515 | No |
| 67 | Ctss | 7002 | -0.065 | 0.0427 | No |
| 68 | Car2 | 7104 | -0.072 | 0.0368 | No |
| 69 | Tfpi | 7118 | -0.073 | 0.0380 | No |
| 70 | Ank | 7180 | -0.077 | 0.0355 | No |
| 71 | Map3k1 | 7447 | -0.100 | 0.0170 | No |
| 72 | Itga2 | 7865 | -0.132 | -0.0128 | No |
| 73 | Traf1 | 7880 | -0.134 | -0.0097 | No |
| 74 | Btbd3 | 7917 | -0.136 | -0.0084 | No |
| 75 | Cbl | 8141 | -0.155 | -0.0217 | No |
| 76 | Dcbld2 | 8364 | -0.173 | -0.0343 | No |
| 77 | Csf2ra | 8417 | -0.176 | -0.0330 | No |
| 78 | Igf2 | 8857 | -0.209 | -0.0622 | No |
| 79 | Ptcd2 | 8979 | -0.217 | -0.0652 | No |
| 80 | Gypc | 9087 | -0.226 | -0.0667 | No |
| 81 | Zfp277 | 9357 | -0.246 | -0.0809 | No |
| 82 | Nin | 9460 | -0.256 | -0.0811 | No |
| 83 | Tnfaip3 | 9668 | -0.274 | -0.0893 | No |
| 84 | Fbxo4 | 9685 | -0.276 | -0.0818 | No |
| 85 | Zfp639 | 9725 | -0.280 | -0.0762 | No |
| 86 | Gprc5b | 9730 | -0.280 | -0.0676 | No |
| 87 | Ikzf1 | 9853 | -0.290 | -0.0684 | No |
| 88 | Ero1a | 10134 | -0.313 | -0.0813 | No |
| 89 | Trib2 | 10170 | -0.316 | -0.0741 | No |
| 90 | Peg3 | 10213 | -0.319 | -0.0674 | No |
| 91 | Spry2 | 10214 | -0.319 | -0.0573 | No |
| 92 | Cdadc1 | 10421 | -0.339 | -0.0634 | No |
| 93 | Psmb8 | 10656 | -0.365 | -0.0709 | No |
| 94 | Yrdc | 10682 | -0.368 | -0.0613 | No |
| 95 | Wdr33 | 10874 | -0.396 | -0.0643 | No |
| 96 | Irf8 | 10940 | -0.406 | -0.0567 | No |
| 97 | Cab39l | 10965 | -0.410 | -0.0457 | No |
| 98 | Dock2 | 11091 | -0.426 | -0.0424 | No |
| 99 | Glrx | 11097 | -0.428 | -0.0292 | No |
| 100 | Gfpt2 | 11236 | -0.449 | -0.0263 | No |
| 101 | Etv5 | 11449 | -0.484 | -0.0282 | No |
| 102 | Fuca1 | 11675 | -0.531 | -0.0297 | No |
| 103 | Dusp6 | 11885 | -0.596 | -0.0279 | No |
| 104 | Ccnd2 | 11944 | -0.617 | -0.0131 | No |
| 105 | Prelid3b | 12152 | -0.721 | -0.0071 | No |
| 106 | Lat2 | 12213 | -0.765 | 0.0122 | No |

