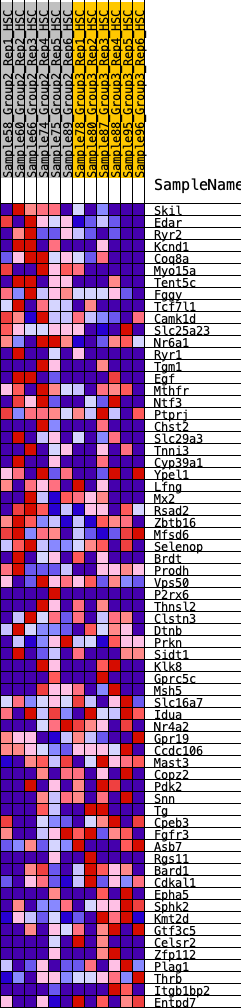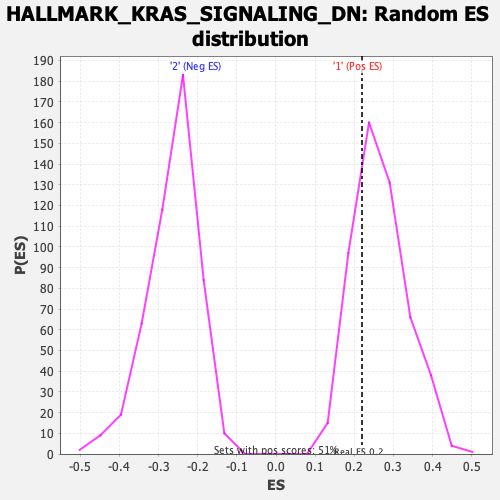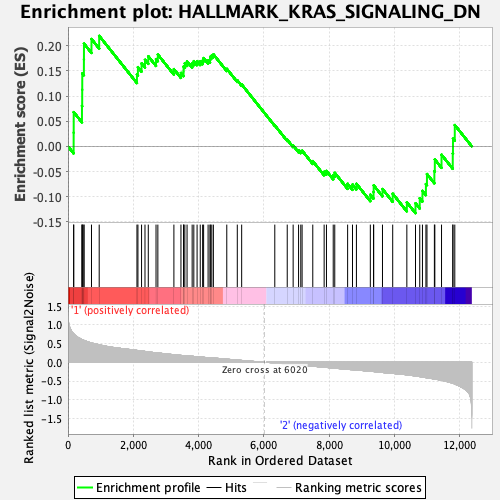
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2197826 |
| Normalized Enrichment Score (NES) | 0.8250377 |
| Nominal p-value | 0.7402344 |
| FDR q-value | 0.95699656 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Skil | 173 | 0.762 | 0.0271 | Yes |
| 2 | Edar | 175 | 0.758 | 0.0681 | Yes |
| 3 | Ryr2 | 428 | 0.607 | 0.0804 | Yes |
| 4 | Kcnd1 | 435 | 0.606 | 0.1127 | Yes |
| 5 | Coq8a | 437 | 0.604 | 0.1452 | Yes |
| 6 | Myo15a | 487 | 0.585 | 0.1729 | Yes |
| 7 | Tent5c | 489 | 0.584 | 0.2044 | Yes |
| 8 | Fggy | 719 | 0.514 | 0.2136 | Yes |
| 9 | Tcf7l1 | 955 | 0.468 | 0.2198 | Yes |
| 10 | Camk1d | 2111 | 0.317 | 0.1430 | No |
| 11 | Slc25a23 | 2141 | 0.313 | 0.1575 | No |
| 12 | Nr6a1 | 2252 | 0.303 | 0.1650 | No |
| 13 | Ryr1 | 2357 | 0.290 | 0.1722 | No |
| 14 | Tgm1 | 2458 | 0.278 | 0.1791 | No |
| 15 | Egf | 2695 | 0.253 | 0.1736 | No |
| 16 | Mthfr | 2751 | 0.248 | 0.1826 | No |
| 17 | Ntf3 | 3242 | 0.202 | 0.1536 | No |
| 18 | Ptprj | 3458 | 0.184 | 0.1461 | No |
| 19 | Chst2 | 3538 | 0.177 | 0.1492 | No |
| 20 | Slc29a3 | 3539 | 0.177 | 0.1588 | No |
| 21 | Tnni3 | 3577 | 0.175 | 0.1652 | No |
| 22 | Cyp39a1 | 3645 | 0.173 | 0.1691 | No |
| 23 | Ypel1 | 3801 | 0.159 | 0.1651 | No |
| 24 | Lfng | 3852 | 0.156 | 0.1695 | No |
| 25 | Mx2 | 3952 | 0.149 | 0.1695 | No |
| 26 | Rsad2 | 4049 | 0.141 | 0.1693 | No |
| 27 | Zbtb16 | 4122 | 0.135 | 0.1708 | No |
| 28 | Mfsd6 | 4152 | 0.133 | 0.1756 | No |
| 29 | Selenop | 4289 | 0.122 | 0.1711 | No |
| 30 | Brdt | 4348 | 0.117 | 0.1728 | No |
| 31 | Prodh | 4357 | 0.116 | 0.1784 | No |
| 32 | Vps50 | 4402 | 0.113 | 0.1809 | No |
| 33 | P2rx6 | 4451 | 0.110 | 0.1830 | No |
| 34 | Thnsl2 | 4861 | 0.082 | 0.1542 | No |
| 35 | Clstn3 | 5183 | 0.059 | 0.1313 | No |
| 36 | Dtnb | 5317 | 0.049 | 0.1232 | No |
| 37 | Prkn | 6331 | -0.017 | 0.0417 | No |
| 38 | Sidt1 | 6712 | -0.045 | 0.0132 | No |
| 39 | Klk8 | 6893 | -0.057 | 0.0016 | No |
| 40 | Gprc5c | 7059 | -0.069 | -0.0080 | No |
| 41 | Msh5 | 7127 | -0.074 | -0.0095 | No |
| 42 | Slc16a7 | 7167 | -0.076 | -0.0085 | No |
| 43 | Idua | 7494 | -0.104 | -0.0294 | No |
| 44 | Nr4a2 | 7842 | -0.131 | -0.0505 | No |
| 45 | Gpr19 | 7913 | -0.136 | -0.0489 | No |
| 46 | Ccdc106 | 8120 | -0.153 | -0.0573 | No |
| 47 | Mast3 | 8167 | -0.156 | -0.0526 | No |
| 48 | Copz2 | 8561 | -0.187 | -0.0745 | No |
| 49 | Pdk2 | 8710 | -0.195 | -0.0760 | No |
| 50 | Snn | 8830 | -0.207 | -0.0745 | No |
| 51 | Tg | 9256 | -0.240 | -0.0961 | No |
| 52 | Cpeb3 | 9354 | -0.246 | -0.0907 | No |
| 53 | Fgfr3 | 9359 | -0.247 | -0.0776 | No |
| 54 | Asb7 | 9628 | -0.269 | -0.0849 | No |
| 55 | Rgs11 | 9941 | -0.297 | -0.0942 | No |
| 56 | Bard1 | 10374 | -0.332 | -0.1114 | No |
| 57 | Cdkal1 | 10639 | -0.363 | -0.1132 | No |
| 58 | Epha5 | 10769 | -0.381 | -0.1031 | No |
| 59 | Sphk2 | 10852 | -0.392 | -0.0886 | No |
| 60 | Kmt2d | 10958 | -0.409 | -0.0750 | No |
| 61 | Gtf3c5 | 10990 | -0.415 | -0.0551 | No |
| 62 | Celsr2 | 11217 | -0.446 | -0.0493 | No |
| 63 | Zfp112 | 11231 | -0.449 | -0.0261 | No |
| 64 | Plag1 | 11436 | -0.482 | -0.0167 | No |
| 65 | Thrb | 11780 | -0.559 | -0.0144 | No |
| 66 | Itgb1bp2 | 11787 | -0.561 | 0.0155 | No |
| 67 | Entpd7 | 11842 | -0.579 | 0.0424 | No |