Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
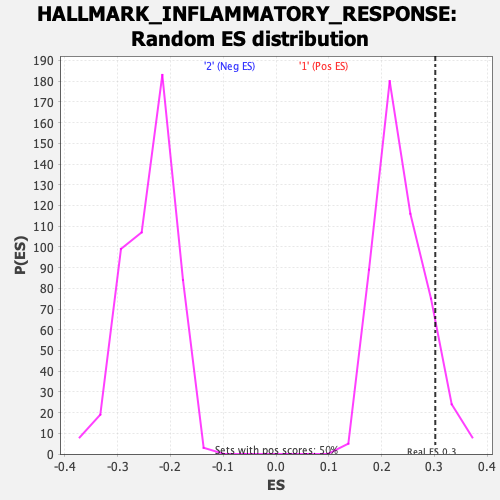
| Enrichment Score (ES) | 0.30161294 |
| Normalized Enrichment Score (NES) | 1.2734797 |
| Nominal p-value | 0.104627766 |
| FDR q-value | 0.3660696 |
| FWER p-Value | 0.815 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 50 | 0.947 | 0.0279 | Yes |
| 2 | Ifitm1 | 87 | 0.871 | 0.0543 | Yes |
| 3 | Osmr | 98 | 0.845 | 0.0820 | Yes |
| 4 | Il4ra | 162 | 0.774 | 0.1029 | Yes |
| 5 | Mxd1 | 185 | 0.751 | 0.1265 | Yes |
| 6 | Irak2 | 201 | 0.737 | 0.1501 | Yes |
| 7 | Ly6e | 400 | 0.617 | 0.1547 | Yes |
| 8 | Itgb3 | 560 | 0.561 | 0.1607 | Yes |
| 9 | Hbegf | 652 | 0.533 | 0.1712 | Yes |
| 10 | Cd82 | 673 | 0.529 | 0.1874 | Yes |
| 11 | Pik3r5 | 851 | 0.488 | 0.1894 | Yes |
| 12 | Nampt | 855 | 0.487 | 0.2056 | Yes |
| 13 | Tlr3 | 1024 | 0.454 | 0.2072 | Yes |
| 14 | Irf1 | 1170 | 0.427 | 0.2098 | Yes |
| 15 | Nfkbia | 1383 | 0.397 | 0.2059 | Yes |
| 16 | Slc31a2 | 1477 | 0.388 | 0.2114 | Yes |
| 17 | Nfkb1 | 1508 | 0.383 | 0.2218 | Yes |
| 18 | Tacr1 | 1566 | 0.377 | 0.2299 | Yes |
| 19 | Il18r1 | 1708 | 0.361 | 0.2306 | Yes |
| 20 | Kif1b | 1745 | 0.359 | 0.2397 | Yes |
| 21 | Ptger4 | 1895 | 0.341 | 0.2391 | Yes |
| 22 | Plaur | 1908 | 0.339 | 0.2495 | Yes |
| 23 | Gch1 | 2022 | 0.328 | 0.2514 | Yes |
| 24 | Abi1 | 2103 | 0.319 | 0.2556 | Yes |
| 25 | Psen1 | 2277 | 0.300 | 0.2516 | Yes |
| 26 | Adrm1 | 2392 | 0.285 | 0.2519 | Yes |
| 27 | Pcdh7 | 2424 | 0.281 | 0.2589 | Yes |
| 28 | Gpr183 | 2444 | 0.279 | 0.2667 | Yes |
| 29 | Emp3 | 2457 | 0.278 | 0.2751 | Yes |
| 30 | Tnfrsf1b | 2506 | 0.272 | 0.2804 | Yes |
| 31 | Axl | 2519 | 0.270 | 0.2885 | Yes |
| 32 | Pde4b | 2521 | 0.270 | 0.2975 | Yes |
| 33 | Ebi3 | 2616 | 0.261 | 0.2987 | Yes |
| 34 | Tnfsf10 | 2739 | 0.250 | 0.2971 | Yes |
| 35 | Scarf1 | 2832 | 0.241 | 0.2978 | Yes |
| 36 | Eif2ak2 | 2883 | 0.236 | 0.3016 | Yes |
| 37 | Il15 | 3037 | 0.221 | 0.2966 | No |
| 38 | Ptafr | 3261 | 0.200 | 0.2851 | No |
| 39 | Nmi | 3279 | 0.198 | 0.2904 | No |
| 40 | Il10ra | 3310 | 0.194 | 0.2945 | No |
| 41 | Rhog | 3477 | 0.182 | 0.2871 | No |
| 42 | Chst2 | 3538 | 0.177 | 0.2882 | No |
| 43 | Rela | 3629 | 0.174 | 0.2867 | No |
| 44 | Klf6 | 3650 | 0.173 | 0.2909 | No |
| 45 | Inhba | 3930 | 0.150 | 0.2732 | No |
| 46 | Lcp2 | 4105 | 0.137 | 0.2636 | No |
| 47 | Ifnar1 | 4287 | 0.122 | 0.2529 | No |
| 48 | Rgs1 | 4418 | 0.112 | 0.2461 | No |
| 49 | Sema4d | 4424 | 0.112 | 0.2495 | No |
| 50 | Slamf1 | 4573 | 0.104 | 0.2409 | No |
| 51 | Il18 | 4716 | 0.094 | 0.2325 | No |
| 52 | Rnf144b | 4799 | 0.087 | 0.2288 | No |
| 53 | Hif1a | 4836 | 0.084 | 0.2287 | No |
| 54 | Acvr1b | 4871 | 0.081 | 0.2286 | No |
| 55 | Atp2c1 | 4908 | 0.079 | 0.2284 | No |
| 56 | Mmp14 | 4916 | 0.078 | 0.2304 | No |
| 57 | Il15ra | 5041 | 0.069 | 0.2227 | No |
| 58 | Csf1 | 5382 | 0.044 | 0.1964 | No |
| 59 | Met | 5620 | 0.027 | 0.1780 | No |
| 60 | Slc31a1 | 5879 | 0.010 | 0.1573 | No |
| 61 | Ccrl2 | 6038 | 0.000 | 0.1444 | No |
| 62 | Slc7a1 | 6272 | -0.013 | 0.1258 | No |
| 63 | Ptpre | 6291 | -0.014 | 0.1248 | No |
| 64 | Icosl | 6302 | -0.015 | 0.1245 | No |
| 65 | Il18rap | 6335 | -0.017 | 0.1225 | No |
| 66 | Tapbp | 6395 | -0.021 | 0.1184 | No |
| 67 | Atp2a2 | 6401 | -0.021 | 0.1187 | No |
| 68 | Ahr | 6567 | -0.034 | 0.1064 | No |
| 69 | Slc28a2 | 6613 | -0.037 | 0.1040 | No |
| 70 | Itga5 | 6986 | -0.064 | 0.0758 | No |
| 71 | P2rx4 | 7014 | -0.066 | 0.0758 | No |
| 72 | Csf3r | 7090 | -0.071 | 0.0721 | No |
| 73 | Gabbr1 | 7393 | -0.096 | 0.0507 | No |
| 74 | Tlr2 | 7441 | -0.100 | 0.0502 | No |
| 75 | Irf7 | 7671 | -0.118 | 0.0355 | No |
| 76 | Icam1 | 7761 | -0.125 | 0.0325 | No |
| 77 | Gpr132 | 7801 | -0.128 | 0.0336 | No |
| 78 | Ripk2 | 8003 | -0.144 | 0.0221 | No |
| 79 | Gnai3 | 8066 | -0.149 | 0.0220 | No |
| 80 | Rtp4 | 8122 | -0.153 | 0.0227 | No |
| 81 | Atp2b1 | 8336 | -0.170 | 0.0111 | No |
| 82 | Dcbld2 | 8364 | -0.173 | 0.0147 | No |
| 83 | Gna15 | 8371 | -0.173 | 0.0200 | No |
| 84 | Sri | 8553 | -0.186 | 0.0115 | No |
| 85 | Myc | 8610 | -0.191 | 0.0134 | No |
| 86 | Acvr2a | 8938 | -0.213 | -0.0061 | No |
| 87 | Nod2 | 8945 | -0.213 | 0.0006 | No |
| 88 | P2ry2 | 9164 | -0.232 | -0.0093 | No |
| 89 | Selenos | 9235 | -0.238 | -0.0070 | No |
| 90 | Lyn | 9410 | -0.251 | -0.0127 | No |
| 91 | Cx3cl1 | 9688 | -0.276 | -0.0260 | No |
| 92 | Lck | 9745 | -0.281 | -0.0211 | No |
| 93 | Pvr | 9976 | -0.298 | -0.0298 | No |
| 94 | Sell | 10039 | -0.305 | -0.0246 | No |
| 95 | Gpc3 | 10282 | -0.325 | -0.0334 | No |
| 96 | Fzd5 | 10331 | -0.329 | -0.0262 | No |
| 97 | Ldlr | 10478 | -0.346 | -0.0265 | No |
| 98 | Il1r1 | 10562 | -0.355 | -0.0213 | No |
| 99 | Calcrl | 10665 | -0.367 | -0.0172 | No |
| 100 | Aqp9 | 10734 | -0.375 | -0.0101 | No |
| 101 | Lpar1 | 11326 | -0.462 | -0.0428 | No |
| 102 | Bst2 | 11805 | -0.566 | -0.0627 | No |
| 103 | P2rx7 | 11827 | -0.573 | -0.0451 | No |
| 104 | Ifngr2 | 12120 | -0.699 | -0.0453 | No |
| 105 | Raf1 | 12131 | -0.707 | -0.0223 | No |
| 106 | Slc11a2 | 12357 | -1.220 | 0.0005 | No |