Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
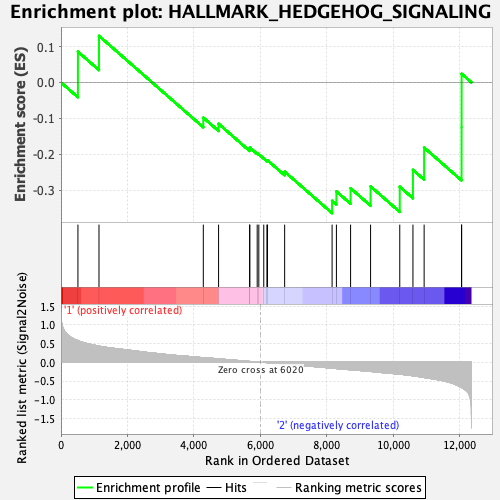
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
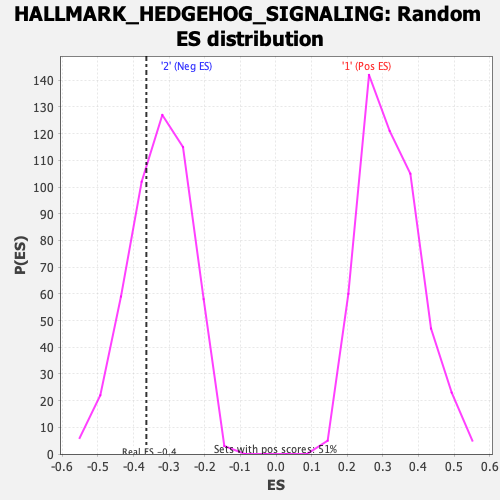
| Enrichment Score (ES) | -0.36392647 |
| Normalized Enrichment Score (NES) | -1.110858 |
| Nominal p-value | 0.3211382 |
| FDR q-value | 0.5957986 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
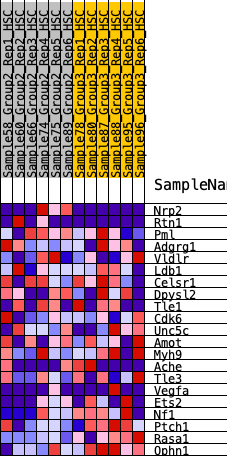
| 1 | Nrp2 | 510 | 0.577 | 0.0856 | No |
| 2 | Rtn1 | 1144 | 0.433 | 0.1296 | No |
| 3 | Pml | 4286 | 0.122 | -0.0980 | No |
| 4 | Adgrg1 | 4746 | 0.091 | -0.1151 | No |
| 5 | Vldlr | 5681 | 0.023 | -0.1858 | No |
| 6 | Ldb1 | 5687 | 0.023 | -0.1812 | No |
| 7 | Celsr1 | 5909 | 0.009 | -0.1972 | No |
| 8 | Dpysl2 | 5958 | 0.005 | -0.2001 | No |
| 9 | Tle1 | 6107 | -0.002 | -0.2116 | No |
| 10 | Cdk6 | 6202 | -0.008 | -0.2175 | No |
| 11 | Unc5c | 6220 | -0.010 | -0.2167 | No |
| 12 | Amot | 6736 | -0.046 | -0.2482 | No |
| 13 | Myh9 | 8165 | -0.156 | -0.3296 | Yes |
| 14 | Ache | 8295 | -0.167 | -0.3033 | Yes |
| 15 | Tle3 | 8722 | -0.197 | -0.2945 | Yes |
| 16 | Vegfa | 9324 | -0.245 | -0.2893 | Yes |
| 17 | Ets2 | 10203 | -0.318 | -0.2904 | Yes |
| 18 | Nf1 | 10600 | -0.359 | -0.2434 | Yes |
| 19 | Ptch1 | 10937 | -0.405 | -0.1814 | Yes |
| 20 | Rasa1 | 12065 | -0.674 | -0.1244 | Yes |
| 21 | Ophn1 | 12066 | -0.675 | 0.0241 | Yes |