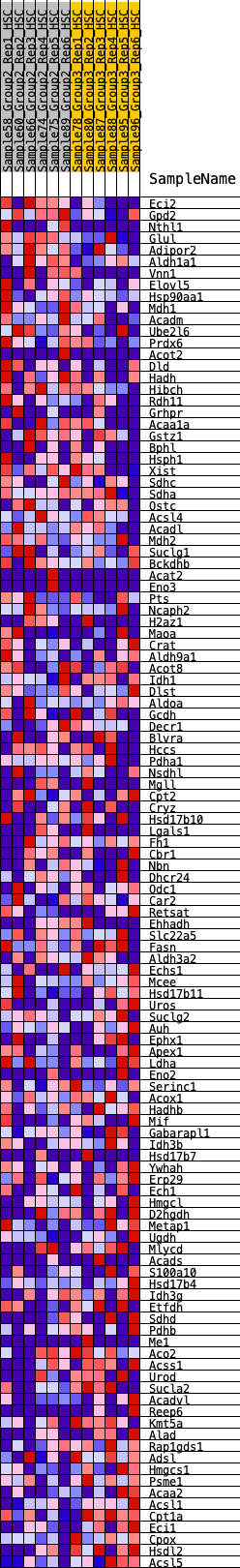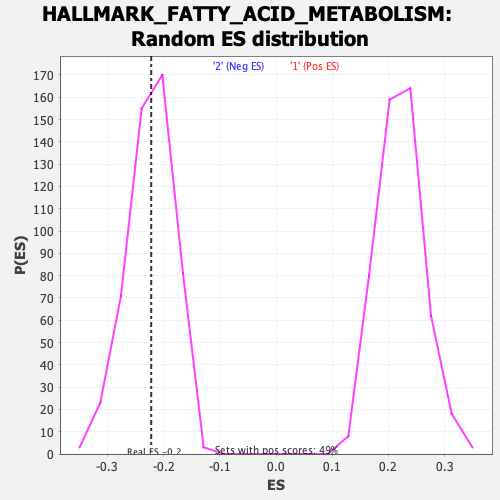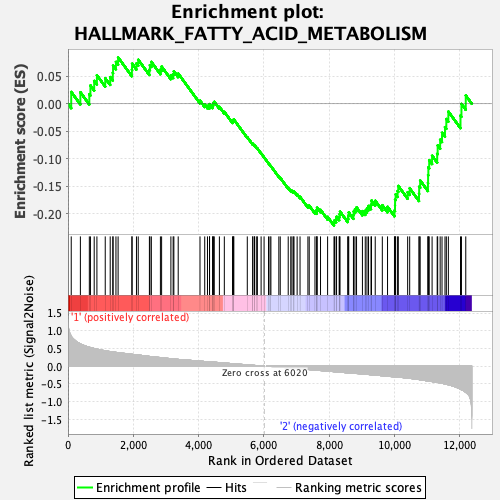
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.22197661 |
| Normalized Enrichment Score (NES) | -0.99374646 |
| Nominal p-value | 0.48221344 |
| FDR q-value | 0.6732948 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eci2 | 99 | 0.841 | 0.0216 | No |
| 2 | Gpd2 | 379 | 0.629 | 0.0210 | No |
| 3 | Nthl1 | 653 | 0.533 | 0.0176 | No |
| 4 | Glul | 687 | 0.526 | 0.0334 | No |
| 5 | Adipor2 | 802 | 0.498 | 0.0417 | No |
| 6 | Aldh1a1 | 885 | 0.482 | 0.0520 | No |
| 7 | Vnn1 | 1140 | 0.433 | 0.0466 | No |
| 8 | Elovl5 | 1292 | 0.409 | 0.0487 | No |
| 9 | Hsp90aa1 | 1372 | 0.399 | 0.0563 | No |
| 10 | Mdh1 | 1380 | 0.397 | 0.0698 | No |
| 11 | Acadm | 1468 | 0.389 | 0.0764 | No |
| 12 | Ube2l6 | 1534 | 0.380 | 0.0845 | No |
| 13 | Prdx6 | 1956 | 0.333 | 0.0619 | No |
| 14 | Acot2 | 1962 | 0.332 | 0.0732 | No |
| 15 | Dld | 2096 | 0.320 | 0.0736 | No |
| 16 | Hadh | 2150 | 0.312 | 0.0803 | No |
| 17 | Hibch | 2492 | 0.274 | 0.0621 | No |
| 18 | Rdh11 | 2508 | 0.272 | 0.0705 | No |
| 19 | Grhpr | 2552 | 0.267 | 0.0764 | No |
| 20 | Acaa1a | 2829 | 0.242 | 0.0624 | No |
| 21 | Gstz1 | 2866 | 0.238 | 0.0678 | No |
| 22 | Bphl | 3150 | 0.210 | 0.0521 | No |
| 23 | Hsph1 | 3219 | 0.204 | 0.0538 | No |
| 24 | Xist | 3241 | 0.202 | 0.0592 | No |
| 25 | Sdhc | 3374 | 0.190 | 0.0551 | No |
| 26 | Sdha | 4041 | 0.142 | 0.0058 | No |
| 27 | Ostc | 4186 | 0.130 | -0.0014 | No |
| 28 | Acsl4 | 4270 | 0.123 | -0.0038 | No |
| 29 | Acadl | 4332 | 0.118 | -0.0046 | No |
| 30 | Mdh2 | 4334 | 0.118 | -0.0006 | No |
| 31 | Suclg1 | 4426 | 0.112 | -0.0040 | No |
| 32 | Bckdhb | 4438 | 0.111 | -0.0010 | No |
| 33 | Acat2 | 4452 | 0.110 | 0.0018 | No |
| 34 | Eno3 | 4478 | 0.110 | 0.0037 | No |
| 35 | Pts | 4635 | 0.100 | -0.0055 | No |
| 36 | Ncaph2 | 4786 | 0.088 | -0.0147 | No |
| 37 | H2az1 | 5037 | 0.069 | -0.0327 | No |
| 38 | Maoa | 5051 | 0.069 | -0.0313 | No |
| 39 | Crat | 5071 | 0.066 | -0.0305 | No |
| 40 | Aldh9a1 | 5074 | 0.066 | -0.0284 | No |
| 41 | Acot8 | 5489 | 0.036 | -0.0609 | No |
| 42 | Idh1 | 5656 | 0.025 | -0.0736 | No |
| 43 | Dlst | 5658 | 0.024 | -0.0728 | No |
| 44 | Aldoa | 5710 | 0.021 | -0.0762 | No |
| 45 | Gcdh | 5716 | 0.020 | -0.0759 | No |
| 46 | Decr1 | 5778 | 0.016 | -0.0803 | No |
| 47 | Blvra | 5797 | 0.016 | -0.0812 | No |
| 48 | Hccs | 5912 | 0.008 | -0.0903 | No |
| 49 | Pdha1 | 6003 | 0.002 | -0.0976 | No |
| 50 | Nsdhl | 6139 | -0.004 | -0.1084 | No |
| 51 | Mgll | 6184 | -0.007 | -0.1118 | No |
| 52 | Cpt2 | 6212 | -0.009 | -0.1137 | No |
| 53 | Cryz | 6455 | -0.025 | -0.1326 | No |
| 54 | Hsd17b10 | 6498 | -0.029 | -0.1350 | No |
| 55 | Lgals1 | 6742 | -0.047 | -0.1532 | No |
| 56 | Fh1 | 6820 | -0.052 | -0.1576 | No |
| 57 | Cbr1 | 6856 | -0.054 | -0.1586 | No |
| 58 | Nbn | 6899 | -0.057 | -0.1600 | No |
| 59 | Dhcr24 | 6919 | -0.059 | -0.1594 | No |
| 60 | Odc1 | 7015 | -0.066 | -0.1649 | No |
| 61 | Car2 | 7104 | -0.072 | -0.1695 | No |
| 62 | Retsat | 7345 | -0.093 | -0.1858 | No |
| 63 | Ehhadh | 7385 | -0.096 | -0.1856 | No |
| 64 | Slc22a5 | 7561 | -0.109 | -0.1961 | No |
| 65 | Fasn | 7616 | -0.113 | -0.1965 | No |
| 66 | Aldh3a2 | 7618 | -0.113 | -0.1926 | No |
| 67 | Echs1 | 7625 | -0.114 | -0.1891 | No |
| 68 | Mcee | 7731 | -0.123 | -0.1933 | No |
| 69 | Hsd17b11 | 7948 | -0.139 | -0.2061 | No |
| 70 | Uros | 8144 | -0.155 | -0.2165 | Yes |
| 71 | Suclg2 | 8161 | -0.156 | -0.2123 | Yes |
| 72 | Auh | 8209 | -0.160 | -0.2105 | Yes |
| 73 | Ephx1 | 8222 | -0.161 | -0.2058 | Yes |
| 74 | Apex1 | 8302 | -0.167 | -0.2063 | Yes |
| 75 | Ldha | 8313 | -0.168 | -0.2012 | Yes |
| 76 | Eno2 | 8328 | -0.169 | -0.1964 | Yes |
| 77 | Serinc1 | 8564 | -0.187 | -0.2090 | Yes |
| 78 | Acox1 | 8581 | -0.188 | -0.2037 | Yes |
| 79 | Hadhb | 8595 | -0.190 | -0.1980 | Yes |
| 80 | Mif | 8739 | -0.198 | -0.2027 | Yes |
| 81 | Gabarapl1 | 8746 | -0.199 | -0.1962 | Yes |
| 82 | Idh3b | 8786 | -0.203 | -0.1922 | Yes |
| 83 | Hsd17b7 | 8834 | -0.207 | -0.1887 | Yes |
| 84 | Ywhah | 9014 | -0.219 | -0.1956 | Yes |
| 85 | Erp29 | 9101 | -0.227 | -0.1946 | Yes |
| 86 | Ech1 | 9153 | -0.232 | -0.1906 | Yes |
| 87 | Hmgcl | 9201 | -0.235 | -0.1861 | Yes |
| 88 | D2hgdh | 9277 | -0.242 | -0.1837 | Yes |
| 89 | Metap1 | 9292 | -0.243 | -0.1763 | Yes |
| 90 | Ugdh | 9406 | -0.251 | -0.1767 | Yes |
| 91 | Mlycd | 9623 | -0.269 | -0.1848 | Yes |
| 92 | Acads | 9783 | -0.285 | -0.1878 | Yes |
| 93 | S100a10 | 9995 | -0.300 | -0.1944 | Yes |
| 94 | Hsd17b4 | 10012 | -0.302 | -0.1851 | Yes |
| 95 | Idh3g | 10014 | -0.302 | -0.1745 | Yes |
| 96 | Etfdh | 10031 | -0.304 | -0.1651 | Yes |
| 97 | Sdhd | 10091 | -0.309 | -0.1590 | Yes |
| 98 | Pdhb | 10112 | -0.310 | -0.1497 | Yes |
| 99 | Me1 | 10399 | -0.336 | -0.1612 | Yes |
| 100 | Aco2 | 10461 | -0.343 | -0.1540 | Yes |
| 101 | Acss1 | 10747 | -0.378 | -0.1639 | Yes |
| 102 | Urod | 10752 | -0.379 | -0.1509 | Yes |
| 103 | Sucla2 | 10782 | -0.383 | -0.1397 | Yes |
| 104 | Acadvl | 11020 | -0.418 | -0.1443 | Yes |
| 105 | Reep6 | 11025 | -0.418 | -0.1299 | Yes |
| 106 | Kmt5a | 11033 | -0.419 | -0.1157 | Yes |
| 107 | Alad | 11059 | -0.422 | -0.1028 | Yes |
| 108 | Rap1gds1 | 11147 | -0.435 | -0.0945 | Yes |
| 109 | Adsl | 11303 | -0.459 | -0.0910 | Yes |
| 110 | Hmgcs1 | 11318 | -0.461 | -0.0759 | Yes |
| 111 | Psme1 | 11394 | -0.473 | -0.0653 | Yes |
| 112 | Acaa2 | 11451 | -0.485 | -0.0528 | Yes |
| 113 | Acsl1 | 11542 | -0.501 | -0.0424 | Yes |
| 114 | Cpt1a | 11583 | -0.510 | -0.0277 | Yes |
| 115 | Eci1 | 11645 | -0.524 | -0.0142 | Yes |
| 116 | Cpox | 12017 | -0.652 | -0.0214 | Yes |
| 117 | Hsdl2 | 12046 | -0.665 | -0.0002 | Yes |
| 118 | Acsl5 | 12178 | -0.737 | 0.0151 | Yes |