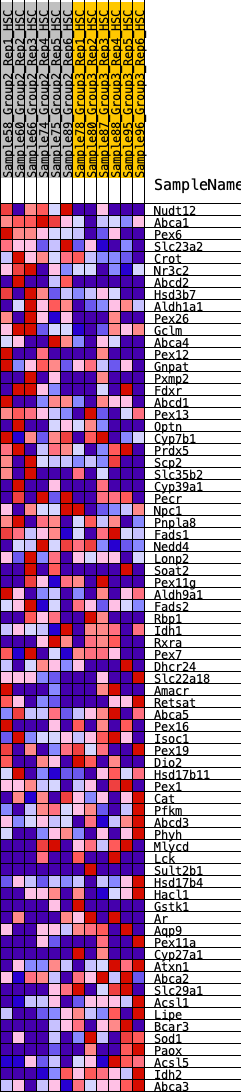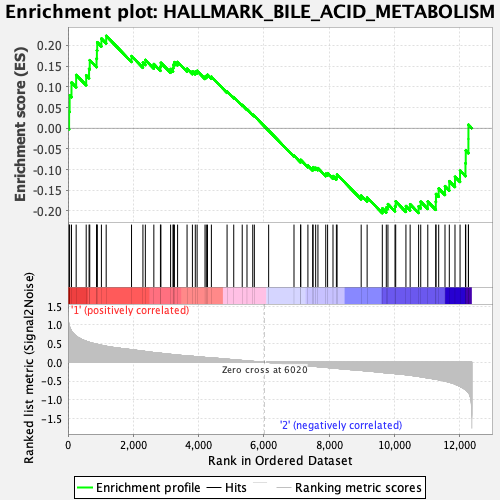
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.2230284 |
| Normalized Enrichment Score (NES) | 0.98920023 |
| Nominal p-value | 0.51028806 |
| FDR q-value | 0.65995115 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nudt12 | 34 | 1.006 | 0.0401 | Yes |
| 2 | Abca1 | 50 | 0.947 | 0.0793 | Yes |
| 3 | Pex6 | 107 | 0.834 | 0.1103 | Yes |
| 4 | Slc23a2 | 250 | 0.700 | 0.1286 | Yes |
| 5 | Crot | 557 | 0.562 | 0.1276 | Yes |
| 6 | Nr3c2 | 645 | 0.534 | 0.1433 | Yes |
| 7 | Abcd2 | 666 | 0.530 | 0.1643 | Yes |
| 8 | Hsd3b7 | 874 | 0.484 | 0.1681 | Yes |
| 9 | Aldh1a1 | 885 | 0.482 | 0.1878 | Yes |
| 10 | Pex26 | 895 | 0.481 | 0.2076 | Yes |
| 11 | Gclm | 1023 | 0.454 | 0.2166 | Yes |
| 12 | Abca4 | 1169 | 0.427 | 0.2230 | Yes |
| 13 | Pex12 | 1945 | 0.334 | 0.1742 | No |
| 14 | Gnpat | 2294 | 0.297 | 0.1586 | No |
| 15 | Pxmp2 | 2368 | 0.288 | 0.1649 | No |
| 16 | Fdxr | 2628 | 0.260 | 0.1549 | No |
| 17 | Abcd1 | 2830 | 0.241 | 0.1488 | No |
| 18 | Pex13 | 2844 | 0.240 | 0.1580 | No |
| 19 | Optn | 3141 | 0.211 | 0.1429 | No |
| 20 | Cyp7b1 | 3216 | 0.204 | 0.1456 | No |
| 21 | Prdx5 | 3227 | 0.203 | 0.1535 | No |
| 22 | Scp2 | 3258 | 0.200 | 0.1596 | No |
| 23 | Slc35b2 | 3355 | 0.192 | 0.1599 | No |
| 24 | Cyp39a1 | 3645 | 0.173 | 0.1438 | No |
| 25 | Pecr | 3809 | 0.159 | 0.1373 | No |
| 26 | Npc1 | 3897 | 0.152 | 0.1367 | No |
| 27 | Pnpla8 | 3953 | 0.149 | 0.1385 | No |
| 28 | Fads1 | 4195 | 0.129 | 0.1244 | No |
| 29 | Nedd4 | 4235 | 0.126 | 0.1267 | No |
| 30 | Lonp2 | 4271 | 0.123 | 0.1291 | No |
| 31 | Soat2 | 4390 | 0.114 | 0.1243 | No |
| 32 | Pex11g | 4870 | 0.081 | 0.0888 | No |
| 33 | Aldh9a1 | 5074 | 0.066 | 0.0751 | No |
| 34 | Fads2 | 5334 | 0.048 | 0.0561 | No |
| 35 | Rbp1 | 5480 | 0.036 | 0.0459 | No |
| 36 | Idh1 | 5656 | 0.025 | 0.0327 | No |
| 37 | Rxra | 5705 | 0.021 | 0.0297 | No |
| 38 | Pex7 | 6143 | -0.004 | -0.0057 | No |
| 39 | Dhcr24 | 6919 | -0.059 | -0.0663 | No |
| 40 | Slc22a18 | 7123 | -0.074 | -0.0797 | No |
| 41 | Amacr | 7124 | -0.074 | -0.0765 | No |
| 42 | Retsat | 7345 | -0.093 | -0.0905 | No |
| 43 | Abca5 | 7492 | -0.104 | -0.0979 | No |
| 44 | Pex16 | 7503 | -0.105 | -0.0942 | No |
| 45 | Isoc1 | 7582 | -0.110 | -0.0959 | No |
| 46 | Pex19 | 7652 | -0.116 | -0.0965 | No |
| 47 | Dio2 | 7891 | -0.134 | -0.1102 | No |
| 48 | Hsd17b11 | 7948 | -0.139 | -0.1088 | No |
| 49 | Pex1 | 8113 | -0.153 | -0.1157 | No |
| 50 | Cat | 8217 | -0.160 | -0.1172 | No |
| 51 | Pfkm | 8238 | -0.162 | -0.1119 | No |
| 52 | Abcd3 | 8976 | -0.216 | -0.1627 | No |
| 53 | Phyh | 9158 | -0.232 | -0.1675 | No |
| 54 | Mlycd | 9623 | -0.269 | -0.1938 | No |
| 55 | Lck | 9745 | -0.281 | -0.1917 | No |
| 56 | Sult2b1 | 9795 | -0.286 | -0.1835 | No |
| 57 | Hsd17b4 | 10012 | -0.302 | -0.1882 | No |
| 58 | Hacl1 | 10033 | -0.304 | -0.1769 | No |
| 59 | Gstk1 | 10346 | -0.331 | -0.1881 | No |
| 60 | Ar | 10475 | -0.345 | -0.1838 | No |
| 61 | Aqp9 | 10734 | -0.375 | -0.1888 | No |
| 62 | Pex11a | 10799 | -0.385 | -0.1776 | No |
| 63 | Cyp27a1 | 11015 | -0.417 | -0.1773 | No |
| 64 | Atxn1 | 11260 | -0.452 | -0.1779 | No |
| 65 | Abca2 | 11266 | -0.453 | -0.1590 | No |
| 66 | Slc29a1 | 11349 | -0.466 | -0.1458 | No |
| 67 | Acsl1 | 11542 | -0.501 | -0.1401 | No |
| 68 | Lipe | 11674 | -0.531 | -0.1281 | No |
| 69 | Bcar3 | 11848 | -0.581 | -0.1174 | No |
| 70 | Sod1 | 12003 | -0.643 | -0.1025 | No |
| 71 | Paox | 12169 | -0.731 | -0.0847 | No |
| 72 | Acsl5 | 12178 | -0.737 | -0.0540 | No |
| 73 | Idh2 | 12259 | -0.807 | -0.0261 | No |
| 74 | Abca3 | 12260 | -0.808 | 0.0084 | No |