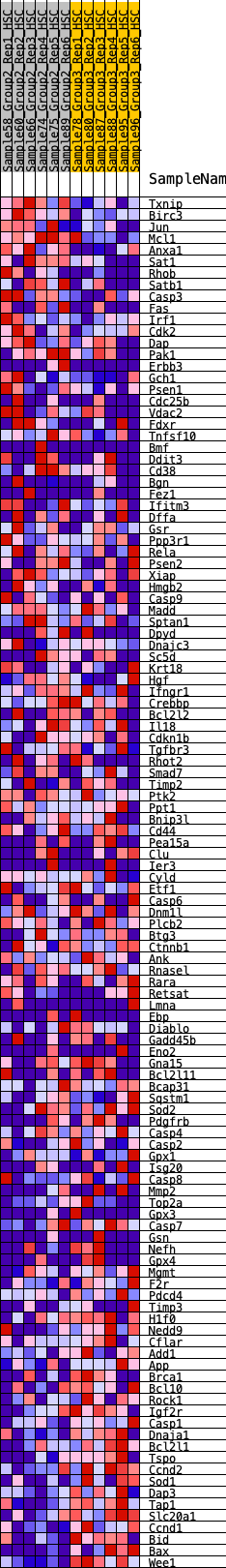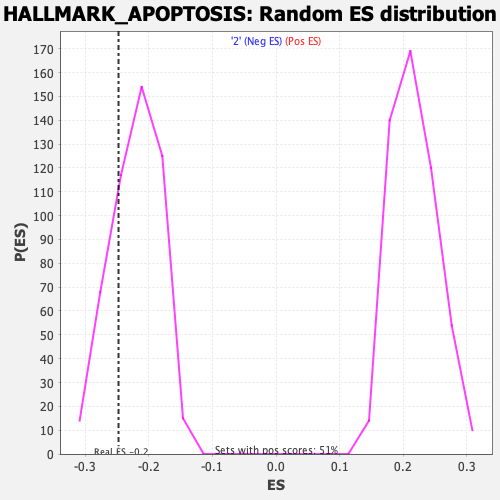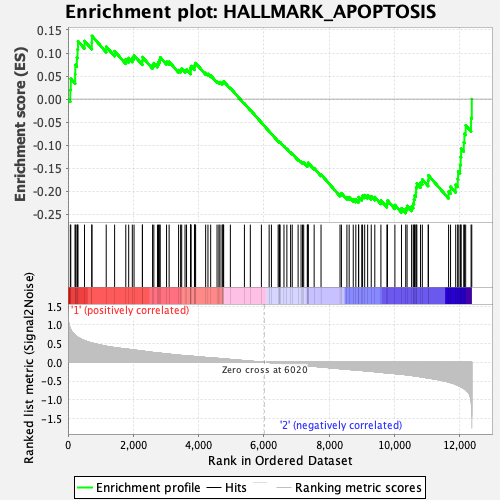
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group2_versus_Group3.HSC_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.24772282 |
| Normalized Enrichment Score (NES) | -1.1210507 |
| Nominal p-value | 0.23123732 |
| FDR q-value | 0.62245905 |
| FWER p-Value | 0.978 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Txnip | 71 | 0.904 | 0.0202 | No |
| 2 | Birc3 | 85 | 0.873 | 0.0442 | No |
| 3 | Jun | 219 | 0.717 | 0.0539 | No |
| 4 | Mcl1 | 223 | 0.715 | 0.0742 | No |
| 5 | Anxa1 | 272 | 0.688 | 0.0901 | No |
| 6 | Sat1 | 292 | 0.672 | 0.1078 | No |
| 7 | Rhob | 306 | 0.661 | 0.1258 | No |
| 8 | Satb1 | 505 | 0.578 | 0.1262 | No |
| 9 | Casp3 | 729 | 0.512 | 0.1227 | No |
| 10 | Fas | 732 | 0.512 | 0.1372 | No |
| 11 | Irf1 | 1170 | 0.427 | 0.1138 | No |
| 12 | Cdk2 | 1427 | 0.392 | 0.1042 | No |
| 13 | Dap | 1769 | 0.355 | 0.0865 | No |
| 14 | Pak1 | 1858 | 0.344 | 0.0892 | No |
| 15 | Erbb3 | 1969 | 0.332 | 0.0898 | No |
| 16 | Gch1 | 2022 | 0.328 | 0.0949 | No |
| 17 | Psen1 | 2277 | 0.300 | 0.0828 | No |
| 18 | Cdc25b | 2279 | 0.300 | 0.0914 | No |
| 19 | Vdac2 | 2589 | 0.263 | 0.0737 | No |
| 20 | Fdxr | 2628 | 0.260 | 0.0780 | No |
| 21 | Tnfsf10 | 2739 | 0.250 | 0.0762 | No |
| 22 | Bmf | 2771 | 0.247 | 0.0808 | No |
| 23 | Ddit3 | 2806 | 0.244 | 0.0850 | No |
| 24 | Cd38 | 2822 | 0.243 | 0.0908 | No |
| 25 | Bgn | 3017 | 0.223 | 0.0813 | No |
| 26 | Fez1 | 3096 | 0.216 | 0.0812 | No |
| 27 | Ifitm3 | 3386 | 0.189 | 0.0630 | No |
| 28 | Dffa | 3448 | 0.184 | 0.0633 | No |
| 29 | Gsr | 3481 | 0.182 | 0.0659 | No |
| 30 | Ppp3r1 | 3583 | 0.174 | 0.0627 | No |
| 31 | Rela | 3629 | 0.174 | 0.0640 | No |
| 32 | Psen2 | 3751 | 0.163 | 0.0588 | No |
| 33 | Xiap | 3752 | 0.163 | 0.0635 | No |
| 34 | Hmgb2 | 3755 | 0.163 | 0.0680 | No |
| 35 | Casp9 | 3767 | 0.162 | 0.0717 | No |
| 36 | Madd | 3872 | 0.154 | 0.0677 | No |
| 37 | Sptan1 | 3873 | 0.153 | 0.0721 | No |
| 38 | Dpyd | 3890 | 0.153 | 0.0751 | No |
| 39 | Dnajc3 | 3907 | 0.151 | 0.0782 | No |
| 40 | Sc5d | 4214 | 0.128 | 0.0569 | No |
| 41 | Krt18 | 4285 | 0.122 | 0.0547 | No |
| 42 | Hgf | 4368 | 0.116 | 0.0513 | No |
| 43 | Ifngr1 | 4563 | 0.105 | 0.0385 | No |
| 44 | Crebbp | 4620 | 0.101 | 0.0368 | No |
| 45 | Bcl2l2 | 4653 | 0.099 | 0.0370 | No |
| 46 | Il18 | 4716 | 0.094 | 0.0347 | No |
| 47 | Cdkn1b | 4739 | 0.092 | 0.0355 | No |
| 48 | Tgfbr3 | 4755 | 0.090 | 0.0369 | No |
| 49 | Rhot2 | 4766 | 0.090 | 0.0386 | No |
| 50 | Smad7 | 4971 | 0.075 | 0.0241 | No |
| 51 | Timp2 | 5401 | 0.042 | -0.0097 | No |
| 52 | Ptk2 | 5582 | 0.030 | -0.0235 | No |
| 53 | Ppt1 | 5921 | 0.008 | -0.0509 | No |
| 54 | Bnip3l | 6158 | -0.005 | -0.0700 | No |
| 55 | Cd44 | 6228 | -0.010 | -0.0754 | No |
| 56 | Pea15a | 6434 | -0.024 | -0.0914 | No |
| 57 | Clu | 6467 | -0.026 | -0.0933 | No |
| 58 | Ier3 | 6474 | -0.027 | -0.0930 | No |
| 59 | Cyld | 6494 | -0.029 | -0.0937 | No |
| 60 | Etf1 | 6612 | -0.037 | -0.1022 | No |
| 61 | Casp6 | 6701 | -0.044 | -0.1081 | No |
| 62 | Dnm1l | 6815 | -0.051 | -0.1159 | No |
| 63 | Plcb2 | 6865 | -0.055 | -0.1183 | No |
| 64 | Btg3 | 7045 | -0.069 | -0.1310 | No |
| 65 | Ctnnb1 | 7135 | -0.074 | -0.1361 | No |
| 66 | Ank | 7180 | -0.077 | -0.1375 | No |
| 67 | Rnasel | 7216 | -0.081 | -0.1380 | No |
| 68 | Rara | 7326 | -0.091 | -0.1443 | No |
| 69 | Retsat | 7345 | -0.093 | -0.1431 | No |
| 70 | Lmna | 7349 | -0.093 | -0.1407 | No |
| 71 | Ebp | 7352 | -0.094 | -0.1382 | No |
| 72 | Diablo | 7536 | -0.107 | -0.1500 | No |
| 73 | Gadd45b | 7747 | -0.124 | -0.1636 | No |
| 74 | Eno2 | 8328 | -0.169 | -0.2061 | No |
| 75 | Gna15 | 8371 | -0.173 | -0.2046 | No |
| 76 | Bcl2l11 | 8539 | -0.185 | -0.2129 | No |
| 77 | Bcap31 | 8608 | -0.191 | -0.2130 | No |
| 78 | Sqstm1 | 8733 | -0.198 | -0.2174 | No |
| 79 | Sod2 | 8812 | -0.205 | -0.2179 | No |
| 80 | Pdgfrb | 8896 | -0.210 | -0.2186 | No |
| 81 | Casp4 | 8899 | -0.211 | -0.2127 | No |
| 82 | Casp2 | 8994 | -0.218 | -0.2142 | No |
| 83 | Gpx1 | 9019 | -0.220 | -0.2098 | No |
| 84 | Isg20 | 9083 | -0.226 | -0.2085 | No |
| 85 | Casp8 | 9174 | -0.233 | -0.2091 | No |
| 86 | Mmp2 | 9284 | -0.242 | -0.2111 | No |
| 87 | Top2a | 9393 | -0.250 | -0.2127 | No |
| 88 | Gpx3 | 9580 | -0.267 | -0.2202 | No |
| 89 | Casp7 | 9764 | -0.284 | -0.2270 | No |
| 90 | Gsn | 9782 | -0.285 | -0.2202 | No |
| 91 | Nefh | 10008 | -0.301 | -0.2299 | No |
| 92 | Gpx4 | 10210 | -0.319 | -0.2372 | No |
| 93 | Mgmt | 10340 | -0.330 | -0.2382 | Yes |
| 94 | F2r | 10381 | -0.333 | -0.2319 | Yes |
| 95 | Pdcd4 | 10520 | -0.350 | -0.2332 | Yes |
| 96 | Timp3 | 10570 | -0.356 | -0.2269 | Yes |
| 97 | H1f0 | 10592 | -0.359 | -0.2183 | Yes |
| 98 | Nedd9 | 10609 | -0.361 | -0.2093 | Yes |
| 99 | Cflar | 10653 | -0.364 | -0.2023 | Yes |
| 100 | Add1 | 10657 | -0.365 | -0.1921 | Yes |
| 101 | App | 10680 | -0.368 | -0.1833 | Yes |
| 102 | Brca1 | 10793 | -0.384 | -0.1814 | Yes |
| 103 | Bcl10 | 10848 | -0.392 | -0.1746 | Yes |
| 104 | Rock1 | 11027 | -0.418 | -0.1771 | Yes |
| 105 | Igf2r | 11035 | -0.419 | -0.1656 | Yes |
| 106 | Casp1 | 11652 | -0.526 | -0.2008 | Yes |
| 107 | Dnaja1 | 11712 | -0.538 | -0.1901 | Yes |
| 108 | Bcl2l1 | 11870 | -0.591 | -0.1860 | Yes |
| 109 | Tspo | 11932 | -0.613 | -0.1734 | Yes |
| 110 | Ccnd2 | 11944 | -0.617 | -0.1566 | Yes |
| 111 | Sod1 | 12003 | -0.643 | -0.1428 | Yes |
| 112 | Dap3 | 12022 | -0.654 | -0.1255 | Yes |
| 113 | Tap1 | 12036 | -0.662 | -0.1075 | Yes |
| 114 | Slc20a1 | 12117 | -0.698 | -0.0940 | Yes |
| 115 | Ccnd1 | 12137 | -0.711 | -0.0751 | Yes |
| 116 | Bid | 12175 | -0.735 | -0.0571 | Yes |
| 117 | Bax | 12341 | -1.030 | -0.0410 | Yes |
| 118 | Wee1 | 12362 | -1.485 | 0.0001 | Yes |