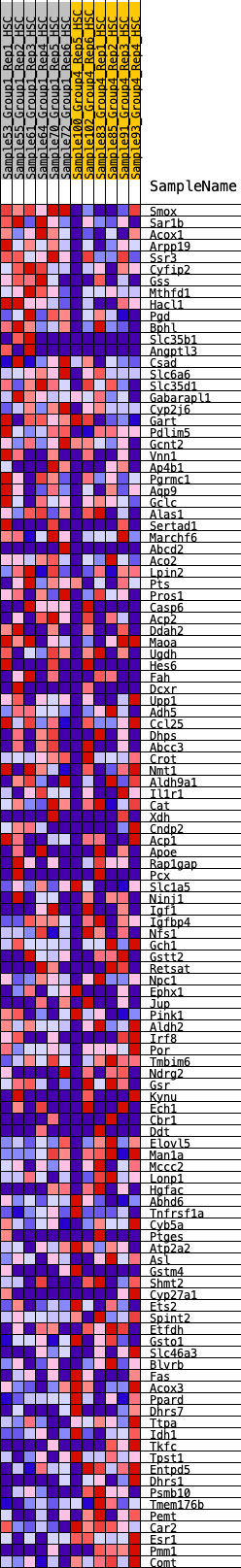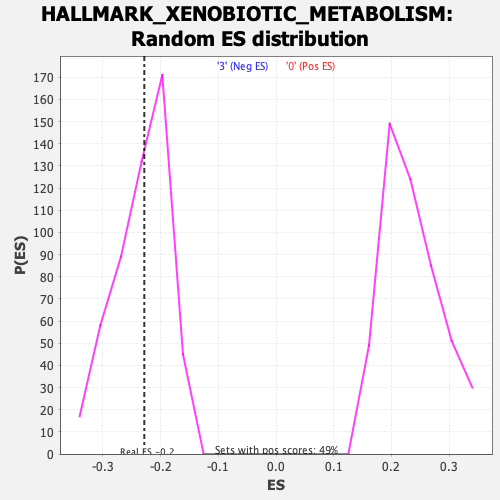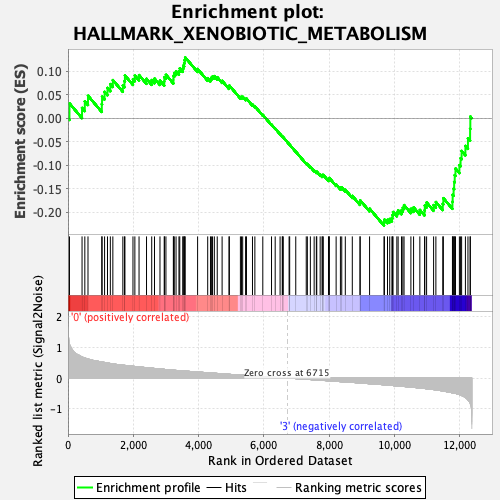
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.22827053 |
| Normalized Enrichment Score (NES) | -0.9828779 |
| Nominal p-value | 0.45703125 |
| FDR q-value | 0.959393 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Smox | 45 | 1.111 | 0.0316 | No |
| 2 | Sar1b | 430 | 0.688 | 0.0220 | No |
| 3 | Acox1 | 515 | 0.654 | 0.0359 | No |
| 4 | Arpp19 | 612 | 0.624 | 0.0479 | No |
| 5 | Ssr3 | 1035 | 0.526 | 0.0301 | No |
| 6 | Cyfip2 | 1043 | 0.525 | 0.0461 | No |
| 7 | Gss | 1120 | 0.510 | 0.0561 | No |
| 8 | Mthfd1 | 1209 | 0.491 | 0.0645 | No |
| 9 | Hacl1 | 1296 | 0.477 | 0.0726 | No |
| 10 | Pgd | 1373 | 0.464 | 0.0811 | No |
| 11 | Bphl | 1679 | 0.422 | 0.0696 | No |
| 12 | Slc35b1 | 1728 | 0.416 | 0.0789 | No |
| 13 | Angptl3 | 1743 | 0.415 | 0.0909 | No |
| 14 | Csad | 1989 | 0.386 | 0.0831 | No |
| 15 | Slc6a6 | 2047 | 0.380 | 0.0905 | No |
| 16 | Slc35d1 | 2175 | 0.366 | 0.0918 | No |
| 17 | Gabarapl1 | 2404 | 0.339 | 0.0839 | No |
| 18 | Cyp2j6 | 2563 | 0.324 | 0.0813 | No |
| 19 | Gart | 2648 | 0.315 | 0.0844 | No |
| 20 | Pdlim5 | 2816 | 0.297 | 0.0802 | No |
| 21 | Gcnt2 | 2948 | 0.285 | 0.0785 | No |
| 22 | Vnn1 | 2951 | 0.285 | 0.0874 | No |
| 23 | Ap4b1 | 2998 | 0.279 | 0.0925 | No |
| 24 | Pgrmc1 | 3227 | 0.257 | 0.0820 | No |
| 25 | Aqp9 | 3228 | 0.257 | 0.0902 | No |
| 26 | Gclc | 3254 | 0.255 | 0.0962 | No |
| 27 | Alas1 | 3308 | 0.251 | 0.0998 | No |
| 28 | Sertad1 | 3397 | 0.244 | 0.1004 | No |
| 29 | Marchf6 | 3422 | 0.242 | 0.1061 | No |
| 30 | Abcd2 | 3514 | 0.233 | 0.1060 | No |
| 31 | Aco2 | 3530 | 0.232 | 0.1122 | No |
| 32 | Lpin2 | 3557 | 0.230 | 0.1173 | No |
| 33 | Pts | 3569 | 0.228 | 0.1237 | No |
| 34 | Pros1 | 3588 | 0.226 | 0.1294 | No |
| 35 | Casp6 | 3968 | 0.201 | 0.1048 | No |
| 36 | Acp2 | 4277 | 0.174 | 0.0851 | No |
| 37 | Ddah2 | 4362 | 0.167 | 0.0836 | No |
| 38 | Maoa | 4390 | 0.164 | 0.0866 | No |
| 39 | Ugdh | 4425 | 0.162 | 0.0890 | No |
| 40 | Hes6 | 4480 | 0.159 | 0.0896 | No |
| 41 | Fah | 4569 | 0.152 | 0.0872 | No |
| 42 | Dcxr | 4718 | 0.140 | 0.0796 | No |
| 43 | Upp1 | 4930 | 0.124 | 0.0663 | No |
| 44 | Adh5 | 4939 | 0.124 | 0.0696 | No |
| 45 | Ccl25 | 5278 | 0.098 | 0.0451 | No |
| 46 | Dhps | 5311 | 0.097 | 0.0456 | No |
| 47 | Abcc3 | 5348 | 0.094 | 0.0456 | No |
| 48 | Crot | 5437 | 0.088 | 0.0412 | No |
| 49 | Nmt1 | 5464 | 0.085 | 0.0417 | No |
| 50 | Aldh9a1 | 5648 | 0.072 | 0.0291 | No |
| 51 | Il1r1 | 5721 | 0.067 | 0.0253 | No |
| 52 | Cat | 5964 | 0.050 | 0.0071 | No |
| 53 | Xdh | 6231 | 0.031 | -0.0136 | No |
| 54 | Cndp2 | 6343 | 0.024 | -0.0219 | No |
| 55 | Acp1 | 6493 | 0.014 | -0.0336 | No |
| 56 | Apoe | 6562 | 0.009 | -0.0389 | No |
| 57 | Rap1gap | 6564 | 0.009 | -0.0387 | No |
| 58 | Pcx | 6591 | 0.007 | -0.0406 | No |
| 59 | Slc1a5 | 6769 | -0.001 | -0.0550 | No |
| 60 | Ninj1 | 6787 | -0.002 | -0.0563 | No |
| 61 | Igf1 | 6973 | -0.014 | -0.0710 | No |
| 62 | Igfbp4 | 7294 | -0.037 | -0.0960 | No |
| 63 | Nfs1 | 7328 | -0.039 | -0.0974 | No |
| 64 | Gch1 | 7420 | -0.046 | -0.1034 | No |
| 65 | Gstt2 | 7536 | -0.054 | -0.1111 | No |
| 66 | Retsat | 7603 | -0.059 | -0.1146 | No |
| 67 | Npc1 | 7616 | -0.059 | -0.1137 | No |
| 68 | Ephx1 | 7720 | -0.068 | -0.1199 | No |
| 69 | Jup | 7782 | -0.072 | -0.1226 | No |
| 70 | Pink1 | 7785 | -0.072 | -0.1205 | No |
| 71 | Aldh2 | 7816 | -0.075 | -0.1206 | No |
| 72 | Irf8 | 7975 | -0.086 | -0.1307 | No |
| 73 | Por | 7994 | -0.088 | -0.1294 | No |
| 74 | Tmbim6 | 7999 | -0.089 | -0.1269 | No |
| 75 | Ndrg2 | 8209 | -0.104 | -0.1407 | No |
| 76 | Gsr | 8340 | -0.114 | -0.1477 | No |
| 77 | Kynu | 8381 | -0.117 | -0.1473 | No |
| 78 | Ech1 | 8490 | -0.125 | -0.1521 | No |
| 79 | Cbr1 | 8704 | -0.140 | -0.1651 | No |
| 80 | Ddt | 8940 | -0.158 | -0.1793 | No |
| 81 | Elovl5 | 8945 | -0.158 | -0.1746 | No |
| 82 | Man1a | 9233 | -0.182 | -0.1923 | No |
| 83 | Mccc2 | 9675 | -0.216 | -0.2214 | Yes |
| 84 | Lonp1 | 9687 | -0.216 | -0.2155 | Yes |
| 85 | Hgfac | 9782 | -0.224 | -0.2160 | Yes |
| 86 | Abhd6 | 9850 | -0.232 | -0.2142 | Yes |
| 87 | Tnfrsf1a | 9917 | -0.238 | -0.2120 | Yes |
| 88 | Cyb5a | 9935 | -0.240 | -0.2058 | Yes |
| 89 | Ptges | 9951 | -0.242 | -0.1993 | Yes |
| 90 | Atp2a2 | 10068 | -0.253 | -0.2008 | Yes |
| 91 | Asl | 10107 | -0.255 | -0.1958 | Yes |
| 92 | Gstm4 | 10210 | -0.266 | -0.1957 | Yes |
| 93 | Shmt2 | 10251 | -0.269 | -0.1904 | Yes |
| 94 | Cyp27a1 | 10288 | -0.273 | -0.1847 | Yes |
| 95 | Ets2 | 10498 | -0.291 | -0.1925 | Yes |
| 96 | Spint2 | 10580 | -0.301 | -0.1896 | Yes |
| 97 | Etfdh | 10772 | -0.323 | -0.1950 | Yes |
| 98 | Gsto1 | 10916 | -0.338 | -0.1959 | Yes |
| 99 | Slc46a3 | 10924 | -0.339 | -0.1857 | Yes |
| 100 | Blvrb | 10979 | -0.347 | -0.1791 | Yes |
| 101 | Fas | 11193 | -0.374 | -0.1847 | Yes |
| 102 | Acox3 | 11263 | -0.384 | -0.1781 | Yes |
| 103 | Ppard | 11474 | -0.418 | -0.1820 | Yes |
| 104 | Dhrs7 | 11492 | -0.423 | -0.1700 | Yes |
| 105 | Ttpa | 11770 | -0.478 | -0.1775 | Yes |
| 106 | Idh1 | 11777 | -0.479 | -0.1628 | Yes |
| 107 | Tkfc | 11806 | -0.488 | -0.1496 | Yes |
| 108 | Tpst1 | 11823 | -0.492 | -0.1353 | Yes |
| 109 | Entpd5 | 11840 | -0.494 | -0.1209 | Yes |
| 110 | Dhrs1 | 11863 | -0.498 | -0.1069 | Yes |
| 111 | Psmb10 | 11983 | -0.537 | -0.0996 | Yes |
| 112 | Tmem176b | 12019 | -0.551 | -0.0850 | Yes |
| 113 | Pemt | 12051 | -0.568 | -0.0695 | Yes |
| 114 | Car2 | 12167 | -0.632 | -0.0589 | Yes |
| 115 | Esr1 | 12245 | -0.711 | -0.0426 | Yes |
| 116 | Pmm1 | 12311 | -0.807 | -0.0223 | Yes |
| 117 | Comt | 12318 | -0.836 | 0.0037 | Yes |