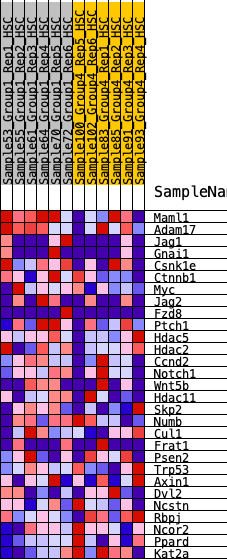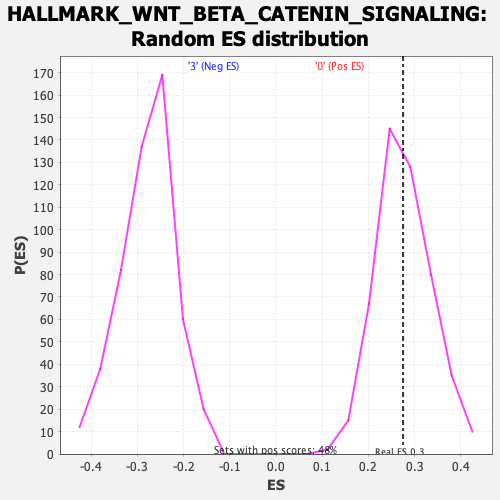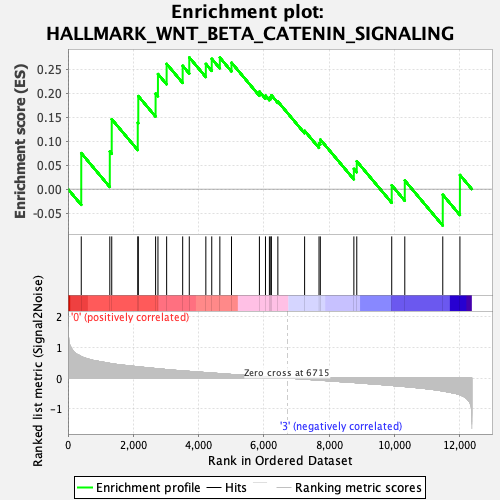
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.27494296 |
| Normalized Enrichment Score (NES) | 0.99441653 |
| Nominal p-value | 0.49585062 |
| FDR q-value | 0.7149729 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Maml1 | 406 | 0.702 | 0.0752 | Yes |
| 2 | Adam17 | 1280 | 0.480 | 0.0784 | Yes |
| 3 | Jag1 | 1339 | 0.470 | 0.1460 | Yes |
| 4 | Gnai1 | 2136 | 0.371 | 0.1386 | Yes |
| 5 | Csnk1e | 2152 | 0.369 | 0.1943 | Yes |
| 6 | Ctnnb1 | 2682 | 0.312 | 0.1994 | Yes |
| 7 | Myc | 2754 | 0.303 | 0.2403 | Yes |
| 8 | Jag2 | 3020 | 0.277 | 0.2614 | Yes |
| 9 | Fzd8 | 3511 | 0.233 | 0.2576 | Yes |
| 10 | Ptch1 | 3713 | 0.219 | 0.2749 | Yes |
| 11 | Hdac5 | 4220 | 0.179 | 0.2615 | No |
| 12 | Hdac2 | 4401 | 0.164 | 0.2721 | No |
| 13 | Ccnd2 | 4649 | 0.146 | 0.2746 | No |
| 14 | Notch1 | 5006 | 0.118 | 0.2639 | No |
| 15 | Wnt5b | 5860 | 0.058 | 0.2037 | No |
| 16 | Hdac11 | 6046 | 0.043 | 0.1953 | No |
| 17 | Skp2 | 6166 | 0.035 | 0.1910 | No |
| 18 | Numb | 6209 | 0.032 | 0.1926 | No |
| 19 | Cul1 | 6226 | 0.031 | 0.1960 | No |
| 20 | Frat1 | 6425 | 0.019 | 0.1829 | No |
| 21 | Psen2 | 7244 | -0.034 | 0.1217 | No |
| 22 | Trp53 | 7685 | -0.065 | 0.0961 | No |
| 23 | Axin1 | 7725 | -0.069 | 0.1035 | No |
| 24 | Dvl2 | 8752 | -0.143 | 0.0423 | No |
| 25 | Ncstn | 8841 | -0.149 | 0.0581 | No |
| 26 | Rbpj | 9911 | -0.238 | 0.0081 | No |
| 27 | Ncor2 | 10311 | -0.275 | 0.0180 | No |
| 28 | Ppard | 11474 | -0.418 | -0.0117 | No |
| 29 | Kat2a | 11999 | -0.543 | 0.0295 | No |