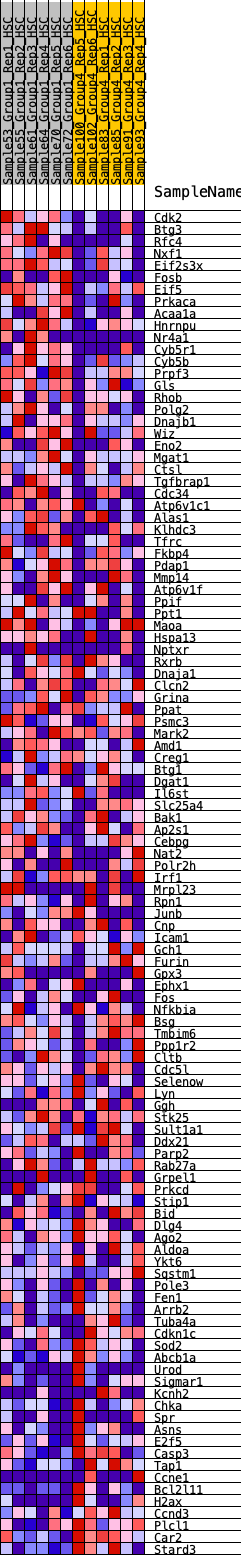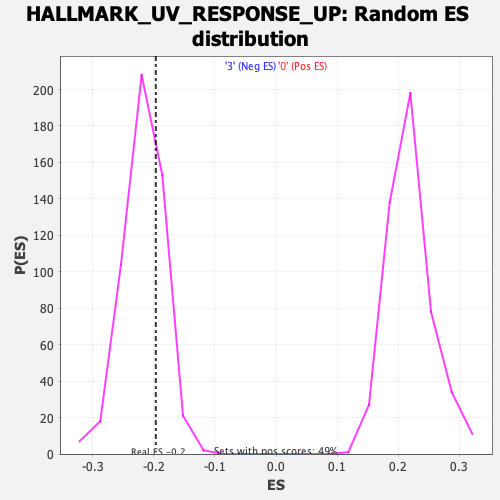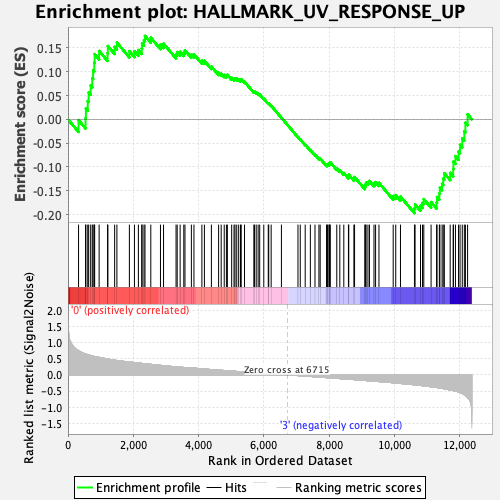
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.19698845 |
| Normalized Enrichment Score (NES) | -0.9032142 |
| Nominal p-value | 0.72319686 |
| FDR q-value | 0.99579954 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdk2 | 323 | 0.751 | -0.0017 | No |
| 2 | Btg3 | 537 | 0.650 | 0.0022 | No |
| 3 | Rfc4 | 548 | 0.645 | 0.0225 | No |
| 4 | Nxf1 | 605 | 0.626 | 0.0385 | No |
| 5 | Eif2s3x | 633 | 0.616 | 0.0565 | No |
| 6 | Fosb | 694 | 0.600 | 0.0712 | No |
| 7 | Eif5 | 743 | 0.587 | 0.0866 | No |
| 8 | Prkaca | 769 | 0.577 | 0.1035 | No |
| 9 | Acaa1a | 805 | 0.570 | 0.1193 | No |
| 10 | Hnrnpu | 816 | 0.567 | 0.1371 | No |
| 11 | Nr4a1 | 953 | 0.541 | 0.1437 | No |
| 12 | Cyb5r1 | 1211 | 0.491 | 0.1388 | No |
| 13 | Cyb5b | 1221 | 0.489 | 0.1541 | No |
| 14 | Prpf3 | 1428 | 0.455 | 0.1523 | No |
| 15 | Gls | 1497 | 0.445 | 0.1613 | No |
| 16 | Rhob | 1879 | 0.400 | 0.1433 | No |
| 17 | Polg2 | 2034 | 0.381 | 0.1432 | No |
| 18 | Dnajb1 | 2155 | 0.369 | 0.1455 | No |
| 19 | Wiz | 2257 | 0.356 | 0.1490 | No |
| 20 | Eno2 | 2268 | 0.355 | 0.1598 | No |
| 21 | Mgat1 | 2326 | 0.349 | 0.1666 | No |
| 22 | Ctsl | 2357 | 0.344 | 0.1754 | No |
| 23 | Tgfbrap1 | 2535 | 0.326 | 0.1717 | No |
| 24 | Cdc34 | 2831 | 0.296 | 0.1573 | No |
| 25 | Atp6v1c1 | 2925 | 0.288 | 0.1591 | No |
| 26 | Alas1 | 3308 | 0.251 | 0.1362 | No |
| 27 | Klhdc3 | 3343 | 0.248 | 0.1415 | No |
| 28 | Tfrc | 3433 | 0.241 | 0.1421 | No |
| 29 | Fkbp4 | 3545 | 0.231 | 0.1407 | No |
| 30 | Pdap1 | 3581 | 0.227 | 0.1452 | No |
| 31 | Mmp14 | 3777 | 0.213 | 0.1363 | No |
| 32 | Atp6v1f | 3856 | 0.208 | 0.1368 | No |
| 33 | Ppif | 4099 | 0.189 | 0.1232 | No |
| 34 | Ppt1 | 4176 | 0.182 | 0.1230 | No |
| 35 | Maoa | 4390 | 0.164 | 0.1110 | No |
| 36 | Hspa13 | 4611 | 0.149 | 0.0979 | No |
| 37 | Nptxr | 4690 | 0.142 | 0.0962 | No |
| 38 | Rxrb | 4783 | 0.137 | 0.0932 | No |
| 39 | Dnaja1 | 4850 | 0.130 | 0.0921 | No |
| 40 | Clcn2 | 4879 | 0.127 | 0.0940 | No |
| 41 | Grina | 5011 | 0.118 | 0.0871 | No |
| 42 | Ppat | 5083 | 0.113 | 0.0851 | No |
| 43 | Psmc3 | 5113 | 0.112 | 0.0864 | No |
| 44 | Mark2 | 5166 | 0.107 | 0.0856 | No |
| 45 | Amd1 | 5225 | 0.102 | 0.0842 | No |
| 46 | Creg1 | 5281 | 0.098 | 0.0830 | No |
| 47 | Btg1 | 5303 | 0.097 | 0.0844 | No |
| 48 | Dgat1 | 5404 | 0.089 | 0.0792 | No |
| 49 | Il6st | 5690 | 0.069 | 0.0582 | No |
| 50 | Slc25a4 | 5711 | 0.068 | 0.0588 | No |
| 51 | Bak1 | 5762 | 0.065 | 0.0569 | No |
| 52 | Ap2s1 | 5821 | 0.060 | 0.0541 | No |
| 53 | Cebpg | 5869 | 0.057 | 0.0521 | No |
| 54 | Nat2 | 5994 | 0.046 | 0.0435 | No |
| 55 | Polr2h | 6131 | 0.037 | 0.0337 | No |
| 56 | Irf1 | 6146 | 0.036 | 0.0337 | No |
| 57 | Mrpl23 | 6220 | 0.031 | 0.0288 | No |
| 58 | Rpn1 | 6534 | 0.011 | 0.0036 | No |
| 59 | Junb | 7041 | -0.019 | -0.0371 | No |
| 60 | Cnp | 7111 | -0.023 | -0.0419 | No |
| 61 | Icam1 | 7262 | -0.035 | -0.0530 | No |
| 62 | Gch1 | 7420 | -0.046 | -0.0643 | No |
| 63 | Furin | 7562 | -0.057 | -0.0740 | No |
| 64 | Gpx3 | 7682 | -0.065 | -0.0816 | No |
| 65 | Ephx1 | 7720 | -0.068 | -0.0824 | No |
| 66 | Fos | 7915 | -0.082 | -0.0955 | No |
| 67 | Nfkbia | 7948 | -0.084 | -0.0954 | No |
| 68 | Bsg | 7960 | -0.084 | -0.0935 | No |
| 69 | Tmbim6 | 7999 | -0.089 | -0.0937 | No |
| 70 | Ppp1r2 | 8007 | -0.089 | -0.0913 | No |
| 71 | Cltb | 8031 | -0.091 | -0.0902 | No |
| 72 | Cdc5l | 8229 | -0.105 | -0.1029 | No |
| 73 | Selenow | 8322 | -0.112 | -0.1067 | No |
| 74 | Lyn | 8444 | -0.121 | -0.1126 | No |
| 75 | Ggh | 8586 | -0.132 | -0.1198 | No |
| 76 | Stk25 | 8596 | -0.132 | -0.1162 | No |
| 77 | Sult1a1 | 8753 | -0.143 | -0.1242 | No |
| 78 | Ddx21 | 8773 | -0.144 | -0.1210 | No |
| 79 | Parp2 | 9086 | -0.169 | -0.1410 | No |
| 80 | Rab27a | 9105 | -0.171 | -0.1368 | No |
| 81 | Grpel1 | 9127 | -0.173 | -0.1329 | No |
| 82 | Prkcd | 9183 | -0.178 | -0.1315 | No |
| 83 | Stip1 | 9229 | -0.182 | -0.1293 | No |
| 84 | Bid | 9368 | -0.192 | -0.1342 | No |
| 85 | Dlg4 | 9417 | -0.196 | -0.1317 | No |
| 86 | Ago2 | 9520 | -0.204 | -0.1333 | No |
| 87 | Aldoa | 9956 | -0.242 | -0.1609 | No |
| 88 | Ykt6 | 10034 | -0.250 | -0.1590 | No |
| 89 | Sqstm1 | 10179 | -0.262 | -0.1621 | No |
| 90 | Pole3 | 10607 | -0.304 | -0.1870 | Yes |
| 91 | Fen1 | 10626 | -0.307 | -0.1784 | Yes |
| 92 | Arrb2 | 10792 | -0.325 | -0.1812 | Yes |
| 93 | Tuba4a | 10854 | -0.331 | -0.1753 | Yes |
| 94 | Cdkn1c | 10893 | -0.336 | -0.1674 | Yes |
| 95 | Sod2 | 11116 | -0.363 | -0.1737 | Yes |
| 96 | Abcb1a | 11285 | -0.387 | -0.1747 | Yes |
| 97 | Urod | 11298 | -0.388 | -0.1629 | Yes |
| 98 | Sigmar1 | 11369 | -0.400 | -0.1555 | Yes |
| 99 | Kcnh2 | 11386 | -0.402 | -0.1437 | Yes |
| 100 | Chka | 11453 | -0.415 | -0.1354 | Yes |
| 101 | Spr | 11488 | -0.422 | -0.1244 | Yes |
| 102 | Asns | 11522 | -0.427 | -0.1131 | Yes |
| 103 | E2f5 | 11701 | -0.464 | -0.1124 | Yes |
| 104 | Casp3 | 11793 | -0.483 | -0.1040 | Yes |
| 105 | Tap1 | 11800 | -0.485 | -0.0886 | Yes |
| 106 | Ccne1 | 11862 | -0.498 | -0.0772 | Yes |
| 107 | Bcl2l11 | 11957 | -0.530 | -0.0675 | Yes |
| 108 | H2ax | 12003 | -0.544 | -0.0533 | Yes |
| 109 | Ccnd3 | 12071 | -0.575 | -0.0399 | Yes |
| 110 | Plcl1 | 12134 | -0.611 | -0.0250 | Yes |
| 111 | Car2 | 12167 | -0.632 | -0.0069 | Yes |
| 112 | Stard3 | 12234 | -0.694 | 0.0105 | Yes |