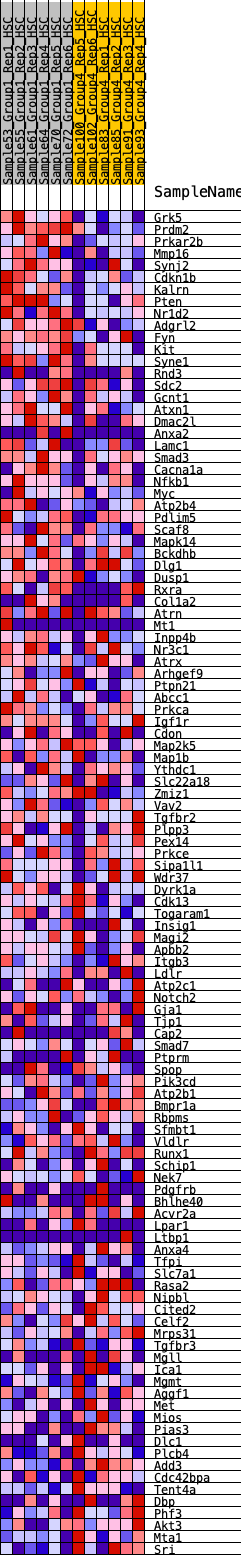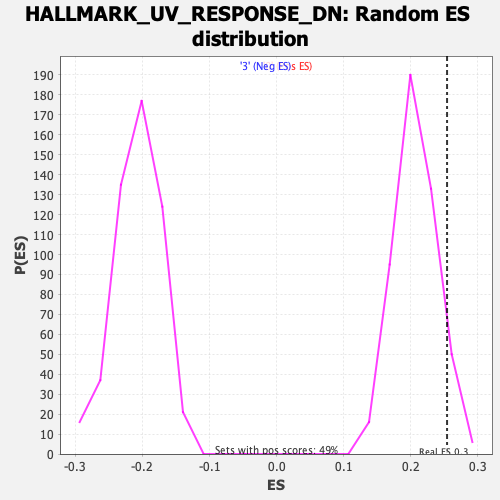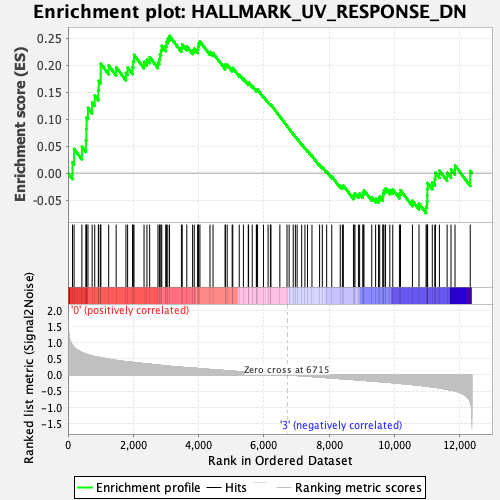
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.2542387 |
| Normalized Enrichment Score (NES) | 1.225204 |
| Nominal p-value | 0.06938776 |
| FDR q-value | 0.5237212 |
| FWER p-Value | 0.895 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Grk5 | 138 | 0.912 | 0.0199 | Yes |
| 2 | Prdm2 | 186 | 0.845 | 0.0448 | Yes |
| 3 | Prkar2b | 425 | 0.690 | 0.0490 | Yes |
| 4 | Mmp16 | 547 | 0.645 | 0.0611 | Yes |
| 5 | Synj2 | 563 | 0.640 | 0.0817 | Yes |
| 6 | Cdkn1b | 569 | 0.638 | 0.1030 | Yes |
| 7 | Kalrn | 611 | 0.624 | 0.1210 | Yes |
| 8 | Pten | 739 | 0.587 | 0.1307 | Yes |
| 9 | Nr1d2 | 818 | 0.567 | 0.1436 | Yes |
| 10 | Adgrl2 | 929 | 0.547 | 0.1533 | Yes |
| 11 | Fyn | 942 | 0.545 | 0.1709 | Yes |
| 12 | Kit | 1001 | 0.534 | 0.1844 | Yes |
| 13 | Syne1 | 1002 | 0.533 | 0.2026 | Yes |
| 14 | Rnd3 | 1245 | 0.485 | 0.1994 | Yes |
| 15 | Sdc2 | 1474 | 0.449 | 0.1961 | Yes |
| 16 | Gcnt1 | 1774 | 0.411 | 0.1857 | Yes |
| 17 | Atxn1 | 1825 | 0.405 | 0.1954 | Yes |
| 18 | Dmac2l | 1975 | 0.388 | 0.1965 | Yes |
| 19 | Anxa2 | 1997 | 0.385 | 0.2079 | Yes |
| 20 | Lamc1 | 2021 | 0.383 | 0.2191 | Yes |
| 21 | Smad3 | 2328 | 0.349 | 0.2060 | Yes |
| 22 | Cacna1a | 2417 | 0.338 | 0.2104 | Yes |
| 23 | Nfkb1 | 2500 | 0.330 | 0.2150 | Yes |
| 24 | Myc | 2754 | 0.303 | 0.2046 | Yes |
| 25 | Atp2b4 | 2797 | 0.298 | 0.2114 | Yes |
| 26 | Pdlim5 | 2816 | 0.297 | 0.2200 | Yes |
| 27 | Scaf8 | 2847 | 0.294 | 0.2276 | Yes |
| 28 | Mapk14 | 2871 | 0.292 | 0.2357 | Yes |
| 29 | Bckdhb | 2992 | 0.280 | 0.2354 | Yes |
| 30 | Dlg1 | 3013 | 0.278 | 0.2433 | Yes |
| 31 | Dusp1 | 3054 | 0.273 | 0.2493 | Yes |
| 32 | Rxra | 3107 | 0.268 | 0.2542 | Yes |
| 33 | Col1a2 | 3472 | 0.236 | 0.2326 | No |
| 34 | Atrn | 3497 | 0.234 | 0.2386 | No |
| 35 | Mt1 | 3637 | 0.224 | 0.2349 | No |
| 36 | Inpp4b | 3815 | 0.211 | 0.2276 | No |
| 37 | Nr3c1 | 3865 | 0.208 | 0.2307 | No |
| 38 | Atrx | 3969 | 0.200 | 0.2292 | No |
| 39 | Arhgef9 | 3984 | 0.199 | 0.2348 | No |
| 40 | Ptpn21 | 4000 | 0.198 | 0.2403 | No |
| 41 | Abcc1 | 4039 | 0.195 | 0.2439 | No |
| 42 | Prkca | 4347 | 0.168 | 0.2245 | No |
| 43 | Igf1r | 4440 | 0.162 | 0.2225 | No |
| 44 | Cdon | 4812 | 0.134 | 0.1968 | No |
| 45 | Map2k5 | 4817 | 0.133 | 0.2010 | No |
| 46 | Map1b | 4874 | 0.128 | 0.2008 | No |
| 47 | Ythdc1 | 5026 | 0.117 | 0.1925 | No |
| 48 | Slc22a18 | 5045 | 0.116 | 0.1950 | No |
| 49 | Zmiz1 | 5242 | 0.101 | 0.1824 | No |
| 50 | Vav2 | 5370 | 0.092 | 0.1752 | No |
| 51 | Tgfbr2 | 5519 | 0.081 | 0.1658 | No |
| 52 | Plpp3 | 5528 | 0.080 | 0.1679 | No |
| 53 | Pex14 | 5640 | 0.072 | 0.1613 | No |
| 54 | Prkce | 5766 | 0.064 | 0.1533 | No |
| 55 | Sipa1l1 | 5771 | 0.064 | 0.1551 | No |
| 56 | Wdr37 | 5804 | 0.061 | 0.1546 | No |
| 57 | Dyrk1a | 5984 | 0.047 | 0.1416 | No |
| 58 | Cdk13 | 6126 | 0.038 | 0.1314 | No |
| 59 | Togaram1 | 6197 | 0.033 | 0.1268 | No |
| 60 | Insig1 | 6214 | 0.032 | 0.1266 | No |
| 61 | Magi2 | 6486 | 0.015 | 0.1050 | No |
| 62 | Apbb2 | 6705 | 0.000 | 0.0872 | No |
| 63 | Itgb3 | 6770 | -0.001 | 0.0820 | No |
| 64 | Ldlr | 6895 | -0.009 | 0.0722 | No |
| 65 | Atp2c1 | 6961 | -0.013 | 0.0673 | No |
| 66 | Notch2 | 7015 | -0.017 | 0.0636 | No |
| 67 | Gja1 | 7155 | -0.027 | 0.0532 | No |
| 68 | Tjp1 | 7254 | -0.034 | 0.0464 | No |
| 69 | Cap2 | 7332 | -0.040 | 0.0414 | No |
| 70 | Smad7 | 7469 | -0.049 | 0.0320 | No |
| 71 | Ptprm | 7703 | -0.067 | 0.0153 | No |
| 72 | Spop | 7786 | -0.073 | 0.0111 | No |
| 73 | Pik3cd | 7919 | -0.082 | 0.0031 | No |
| 74 | Atp2b1 | 8075 | -0.095 | -0.0063 | No |
| 75 | Bmpr1a | 8335 | -0.113 | -0.0236 | No |
| 76 | Rbpms | 8401 | -0.118 | -0.0249 | No |
| 77 | Sfmbt1 | 8430 | -0.120 | -0.0231 | No |
| 78 | Vldlr | 8739 | -0.142 | -0.0434 | No |
| 79 | Runx1 | 8757 | -0.143 | -0.0399 | No |
| 80 | Schip1 | 8786 | -0.145 | -0.0372 | No |
| 81 | Nek7 | 8892 | -0.153 | -0.0406 | No |
| 82 | Pdgfrb | 8928 | -0.156 | -0.0381 | No |
| 83 | Bhlhe40 | 9026 | -0.165 | -0.0404 | No |
| 84 | Acvr2a | 9039 | -0.166 | -0.0357 | No |
| 85 | Lpar1 | 9062 | -0.168 | -0.0318 | No |
| 86 | Ltbp1 | 9300 | -0.188 | -0.0447 | No |
| 87 | Anxa4 | 9418 | -0.196 | -0.0476 | No |
| 88 | Tfpi | 9508 | -0.203 | -0.0479 | No |
| 89 | Slc7a1 | 9543 | -0.206 | -0.0436 | No |
| 90 | Rasa2 | 9640 | -0.213 | -0.0442 | No |
| 91 | Nipbl | 9647 | -0.213 | -0.0374 | No |
| 92 | Cited2 | 9673 | -0.216 | -0.0321 | No |
| 93 | Celf2 | 9722 | -0.220 | -0.0285 | No |
| 94 | Mrps31 | 9853 | -0.232 | -0.0312 | No |
| 95 | Tgfbr3 | 9943 | -0.241 | -0.0303 | No |
| 96 | Mgll | 10147 | -0.260 | -0.0380 | No |
| 97 | Ica1 | 10180 | -0.262 | -0.0316 | No |
| 98 | Mgmt | 10545 | -0.297 | -0.0512 | No |
| 99 | Aggf1 | 10746 | -0.320 | -0.0566 | No |
| 100 | Met | 10963 | -0.344 | -0.0625 | No |
| 101 | Mios | 10992 | -0.348 | -0.0529 | No |
| 102 | Pias3 | 10998 | -0.349 | -0.0414 | No |
| 103 | Dlc1 | 11002 | -0.350 | -0.0297 | No |
| 104 | Plcb4 | 11010 | -0.350 | -0.0183 | No |
| 105 | Add3 | 11153 | -0.367 | -0.0174 | No |
| 106 | Cdc42bpa | 11230 | -0.379 | -0.0107 | No |
| 107 | Tent4a | 11249 | -0.382 | 0.0009 | No |
| 108 | Dbp | 11371 | -0.400 | 0.0047 | No |
| 109 | Phf3 | 11607 | -0.445 | 0.0007 | No |
| 110 | Akt3 | 11726 | -0.470 | 0.0070 | No |
| 111 | Mta1 | 11849 | -0.495 | 0.0140 | No |
| 112 | Sri | 12315 | -0.818 | 0.0039 | No |