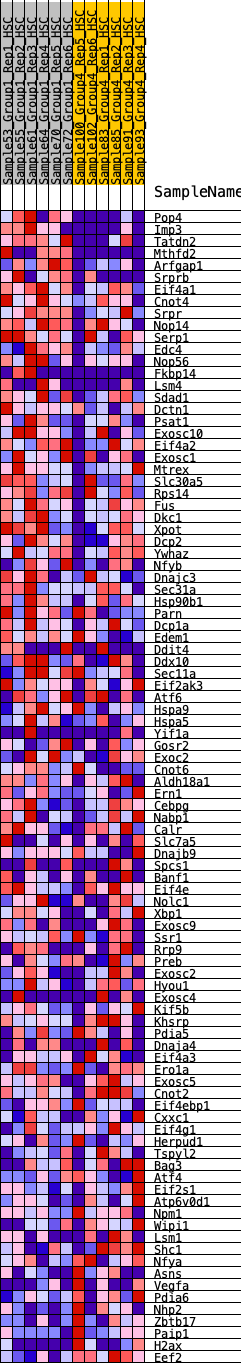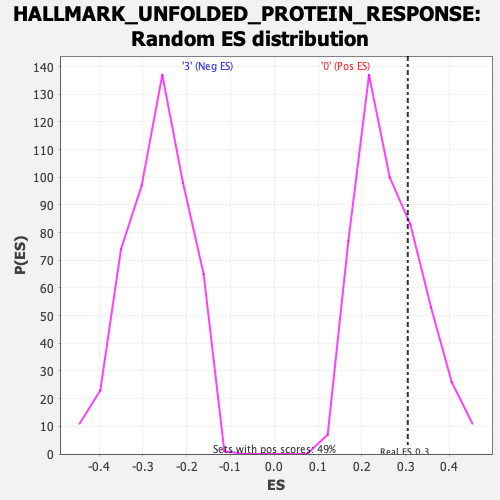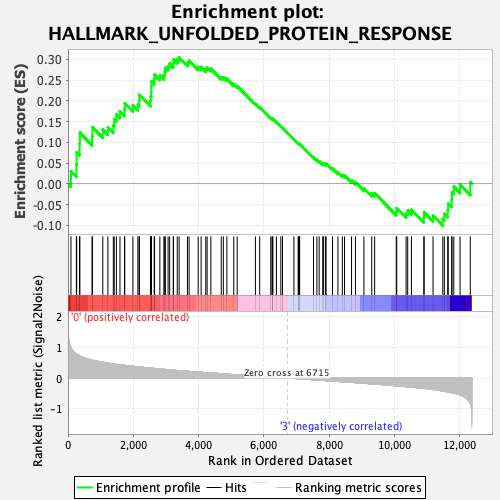
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.30485272 |
| Normalized Enrichment Score (NES) | 1.1492131 |
| Nominal p-value | 0.27732792 |
| FDR q-value | 0.51970595 |
| FWER p-Value | 0.963 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pop4 | 90 | 0.995 | 0.0304 | Yes |
| 2 | Imp3 | 256 | 0.785 | 0.0467 | Yes |
| 3 | Tatdn2 | 265 | 0.783 | 0.0758 | Yes |
| 4 | Mthfd2 | 354 | 0.733 | 0.0964 | Yes |
| 5 | Arfgap1 | 363 | 0.724 | 0.1232 | Yes |
| 6 | Srprb | 737 | 0.587 | 0.1151 | Yes |
| 7 | Eif4a1 | 747 | 0.586 | 0.1366 | Yes |
| 8 | Cnot4 | 1065 | 0.521 | 0.1305 | Yes |
| 9 | Srpr | 1222 | 0.489 | 0.1363 | Yes |
| 10 | Nop14 | 1387 | 0.462 | 0.1405 | Yes |
| 11 | Serp1 | 1415 | 0.459 | 0.1557 | Yes |
| 12 | Edc4 | 1481 | 0.447 | 0.1673 | Yes |
| 13 | Nop56 | 1591 | 0.432 | 0.1749 | Yes |
| 14 | Fkbp14 | 1730 | 0.416 | 0.1794 | Yes |
| 15 | Lsm4 | 1738 | 0.416 | 0.1946 | Yes |
| 16 | Sdad1 | 1987 | 0.387 | 0.1890 | Yes |
| 17 | Dctn1 | 2139 | 0.370 | 0.1908 | Yes |
| 18 | Psat1 | 2181 | 0.365 | 0.2013 | Yes |
| 19 | Exosc10 | 2184 | 0.364 | 0.2150 | Yes |
| 20 | Eif4a2 | 2525 | 0.327 | 0.1997 | Yes |
| 21 | Exosc1 | 2534 | 0.326 | 0.2114 | Yes |
| 22 | Mtrex | 2549 | 0.325 | 0.2226 | Yes |
| 23 | Slc30a5 | 2553 | 0.324 | 0.2346 | Yes |
| 24 | Rps14 | 2556 | 0.324 | 0.2468 | Yes |
| 25 | Fus | 2640 | 0.316 | 0.2520 | Yes |
| 26 | Dkc1 | 2650 | 0.315 | 0.2632 | Yes |
| 27 | Xpot | 2812 | 0.297 | 0.2613 | Yes |
| 28 | Dcp2 | 2935 | 0.287 | 0.2622 | Yes |
| 29 | Ywhaz | 2963 | 0.283 | 0.2708 | Yes |
| 30 | Nfyb | 2981 | 0.281 | 0.2801 | Yes |
| 31 | Dnajc3 | 3064 | 0.272 | 0.2837 | Yes |
| 32 | Sec31a | 3112 | 0.267 | 0.2900 | Yes |
| 33 | Hsp90b1 | 3222 | 0.257 | 0.2909 | Yes |
| 34 | Parn | 3230 | 0.257 | 0.3001 | Yes |
| 35 | Dcp1a | 3345 | 0.248 | 0.3002 | Yes |
| 36 | Edem1 | 3402 | 0.244 | 0.3049 | Yes |
| 37 | Ddit4 | 3657 | 0.223 | 0.2926 | No |
| 38 | Ddx10 | 3708 | 0.219 | 0.2969 | No |
| 39 | Sec11a | 3985 | 0.199 | 0.2819 | No |
| 40 | Eif2ak3 | 4076 | 0.192 | 0.2819 | No |
| 41 | Atf6 | 4218 | 0.179 | 0.2771 | No |
| 42 | Hspa9 | 4259 | 0.175 | 0.2805 | No |
| 43 | Hspa5 | 4374 | 0.166 | 0.2775 | No |
| 44 | Yif1a | 4692 | 0.142 | 0.2571 | No |
| 45 | Gosr2 | 4756 | 0.138 | 0.2572 | No |
| 46 | Exoc2 | 4864 | 0.129 | 0.2533 | No |
| 47 | Cnot6 | 5075 | 0.114 | 0.2406 | No |
| 48 | Aldh18a1 | 5180 | 0.106 | 0.2361 | No |
| 49 | Ern1 | 5738 | 0.066 | 0.1932 | No |
| 50 | Cebpg | 5869 | 0.057 | 0.1848 | No |
| 51 | Nabp1 | 6207 | 0.032 | 0.1585 | No |
| 52 | Calr | 6250 | 0.031 | 0.1563 | No |
| 53 | Slc7a5 | 6254 | 0.030 | 0.1572 | No |
| 54 | Dnajb9 | 6273 | 0.029 | 0.1568 | No |
| 55 | Spcs1 | 6380 | 0.021 | 0.1490 | No |
| 56 | Banf1 | 6516 | 0.012 | 0.1384 | No |
| 57 | Eif4e | 6566 | 0.009 | 0.1348 | No |
| 58 | Nolc1 | 6567 | 0.008 | 0.1351 | No |
| 59 | Xbp1 | 6917 | -0.010 | 0.1070 | No |
| 60 | Exosc9 | 7046 | -0.019 | 0.0973 | No |
| 61 | Ssr1 | 7059 | -0.020 | 0.0971 | No |
| 62 | Rrp9 | 7084 | -0.021 | 0.0960 | No |
| 63 | Preb | 7092 | -0.022 | 0.0962 | No |
| 64 | Exosc2 | 7516 | -0.053 | 0.0637 | No |
| 65 | Hyou1 | 7619 | -0.059 | 0.0577 | No |
| 66 | Exosc4 | 7689 | -0.066 | 0.0546 | No |
| 67 | Kif5b | 7799 | -0.074 | 0.0485 | No |
| 68 | Khsrp | 7826 | -0.076 | 0.0492 | No |
| 69 | Pdia5 | 7888 | -0.080 | 0.0473 | No |
| 70 | Dnaja4 | 7899 | -0.080 | 0.0495 | No |
| 71 | Eif4a3 | 8098 | -0.096 | 0.0370 | No |
| 72 | Ero1a | 8264 | -0.108 | 0.0277 | No |
| 73 | Exosc5 | 8399 | -0.118 | 0.0212 | No |
| 74 | Cnot2 | 8466 | -0.123 | 0.0205 | No |
| 75 | Eif4ebp1 | 8681 | -0.138 | 0.0083 | No |
| 76 | Cxxc1 | 8800 | -0.146 | 0.0042 | No |
| 77 | Eif4g1 | 9060 | -0.167 | -0.0106 | No |
| 78 | Herpud1 | 9299 | -0.188 | -0.0228 | No |
| 79 | Tspyl2 | 9388 | -0.194 | -0.0226 | No |
| 80 | Bag3 | 10050 | -0.252 | -0.0670 | No |
| 81 | Atf4 | 10064 | -0.253 | -0.0584 | No |
| 82 | Eif2s1 | 10352 | -0.279 | -0.0712 | No |
| 83 | Atp6v0d1 | 10400 | -0.283 | -0.0643 | No |
| 84 | Npm1 | 10515 | -0.293 | -0.0625 | No |
| 85 | Wipi1 | 10894 | -0.336 | -0.0806 | No |
| 86 | Lsm1 | 10903 | -0.337 | -0.0685 | No |
| 87 | Shc1 | 11176 | -0.372 | -0.0765 | No |
| 88 | Nfya | 11475 | -0.419 | -0.0849 | No |
| 89 | Asns | 11522 | -0.427 | -0.0725 | No |
| 90 | Vegfa | 11626 | -0.448 | -0.0639 | No |
| 91 | Pdia6 | 11640 | -0.452 | -0.0478 | No |
| 92 | Nhp2 | 11746 | -0.474 | -0.0384 | No |
| 93 | Zbtb17 | 11756 | -0.476 | -0.0211 | No |
| 94 | Paip1 | 11811 | -0.489 | -0.0069 | No |
| 95 | H2ax | 12003 | -0.544 | -0.0019 | No |
| 96 | Eef2 | 12316 | -0.821 | 0.0038 | No |