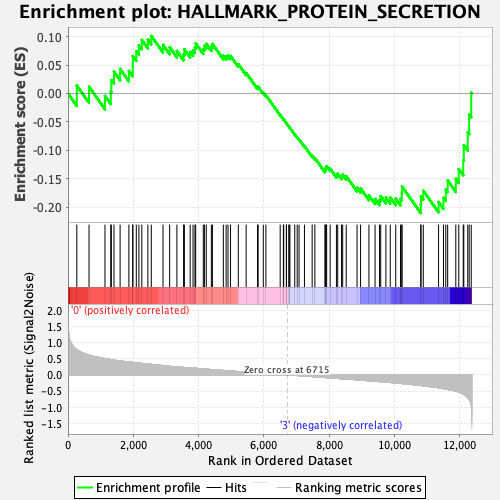
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.2106841 |
| Normalized Enrichment Score (NES) | -0.89533705 |
| Nominal p-value | 0.63380283 |
| FDR q-value | 0.9530528 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cog2 | 270 | 0.778 | 0.0139 | No |
| 2 | Rab2a | 644 | 0.614 | 0.0119 | No |
| 3 | Dop1a | 1132 | 0.508 | -0.0043 | No |
| 4 | Arfgef1 | 1309 | 0.476 | 0.0033 | No |
| 5 | Rab22a | 1327 | 0.472 | 0.0237 | No |
| 6 | Copb1 | 1408 | 0.459 | 0.0384 | No |
| 7 | Arfgap3 | 1595 | 0.432 | 0.0431 | No |
| 8 | Ap2m1 | 1864 | 0.401 | 0.0398 | No |
| 9 | Tpd52 | 1983 | 0.387 | 0.0481 | No |
| 10 | Adam10 | 1985 | 0.387 | 0.0659 | No |
| 11 | Cav2 | 2091 | 0.375 | 0.0746 | No |
| 12 | Tsg101 | 2171 | 0.366 | 0.0851 | No |
| 13 | Cln5 | 2260 | 0.355 | 0.0943 | No |
| 14 | Sec24d | 2446 | 0.336 | 0.0948 | No |
| 15 | Ocrl | 2550 | 0.325 | 0.1014 | No |
| 16 | Rer1 | 2909 | 0.289 | 0.0855 | No |
| 17 | Sec31a | 3112 | 0.267 | 0.0814 | No |
| 18 | Yipf6 | 3334 | 0.248 | 0.0749 | No |
| 19 | Vps4b | 3539 | 0.231 | 0.0689 | No |
| 20 | Ap3b1 | 3563 | 0.229 | 0.0776 | No |
| 21 | Atp6v1h | 3740 | 0.217 | 0.0733 | No |
| 22 | Abca1 | 3828 | 0.210 | 0.0759 | No |
| 23 | Snx2 | 3887 | 0.206 | 0.0807 | No |
| 24 | Cltc | 3909 | 0.204 | 0.0884 | No |
| 25 | Vps45 | 4142 | 0.185 | 0.0780 | No |
| 26 | Ppt1 | 4176 | 0.182 | 0.0837 | No |
| 27 | Mapk1 | 4233 | 0.177 | 0.0874 | No |
| 28 | Ap1g1 | 4388 | 0.165 | 0.0824 | No |
| 29 | Rab5a | 4422 | 0.163 | 0.0872 | No |
| 30 | Gosr2 | 4756 | 0.138 | 0.0665 | No |
| 31 | Dst | 4841 | 0.131 | 0.0657 | No |
| 32 | Arf1 | 4899 | 0.126 | 0.0669 | No |
| 33 | Scamp1 | 4976 | 0.120 | 0.0662 | No |
| 34 | Atp1a1 | 5214 | 0.103 | 0.0517 | No |
| 35 | Ap3s1 | 5454 | 0.086 | 0.0362 | No |
| 36 | Igf2r | 5802 | 0.061 | 0.0107 | No |
| 37 | Ap2s1 | 5821 | 0.060 | 0.0121 | No |
| 38 | Sod1 | 5979 | 0.048 | 0.0015 | No |
| 39 | Zw10 | 6059 | 0.042 | -0.0030 | No |
| 40 | Arfip1 | 6496 | 0.014 | -0.0379 | No |
| 41 | Uso1 | 6594 | 0.007 | -0.0455 | No |
| 42 | Cope | 6604 | 0.006 | -0.0459 | No |
| 43 | Vamp3 | 6686 | 0.002 | -0.0525 | No |
| 44 | Arcn1 | 6688 | 0.002 | -0.0525 | No |
| 45 | Napa | 6764 | -0.001 | -0.0586 | No |
| 46 | Vamp4 | 6768 | -0.001 | -0.0588 | No |
| 47 | Ap2b1 | 6794 | -0.003 | -0.0607 | No |
| 48 | Stx12 | 6942 | -0.012 | -0.0721 | No |
| 49 | Tmed10 | 7017 | -0.018 | -0.0773 | No |
| 50 | Stx7 | 7074 | -0.021 | -0.0809 | No |
| 51 | Stam | 7239 | -0.033 | -0.0927 | No |
| 52 | Gbf1 | 7475 | -0.050 | -0.1096 | No |
| 53 | Dnm1l | 7558 | -0.056 | -0.1137 | No |
| 54 | Lman1 | 7865 | -0.078 | -0.1350 | No |
| 55 | Copb2 | 7890 | -0.080 | -0.1332 | No |
| 56 | Mon2 | 7896 | -0.080 | -0.1299 | No |
| 57 | Sgms1 | 7918 | -0.082 | -0.1279 | No |
| 58 | Rab9 | 8026 | -0.090 | -0.1324 | No |
| 59 | Golga4 | 8218 | -0.105 | -0.1432 | No |
| 60 | Lamp2 | 8258 | -0.107 | -0.1414 | No |
| 61 | Ctsc | 8375 | -0.116 | -0.1455 | No |
| 62 | Rps6ka3 | 8410 | -0.119 | -0.1428 | No |
| 63 | Sec22b | 8517 | -0.127 | -0.1455 | No |
| 64 | Scrn1 | 8849 | -0.149 | -0.1656 | No |
| 65 | Stx16 | 8957 | -0.159 | -0.1670 | No |
| 66 | Ergic3 | 9214 | -0.180 | -0.1795 | No |
| 67 | Bet1 | 9401 | -0.195 | -0.1857 | No |
| 68 | Napg | 9538 | -0.206 | -0.1873 | No |
| 69 | Tmx1 | 9574 | -0.208 | -0.1805 | No |
| 70 | Kif1b | 9733 | -0.221 | -0.1832 | No |
| 71 | Anp32e | 9866 | -0.233 | -0.1832 | No |
| 72 | Ykt6 | 10034 | -0.250 | -0.1853 | No |
| 73 | Ica1 | 10180 | -0.262 | -0.1850 | No |
| 74 | Gnas | 10220 | -0.266 | -0.1758 | No |
| 75 | Galc | 10226 | -0.267 | -0.1639 | No |
| 76 | Clcn3 | 10801 | -0.326 | -0.1956 | Yes |
| 77 | Pam | 10807 | -0.327 | -0.1810 | Yes |
| 78 | M6pr | 10877 | -0.333 | -0.1712 | Yes |
| 79 | Rab14 | 11345 | -0.396 | -0.1910 | Yes |
| 80 | Atp7a | 11495 | -0.423 | -0.1836 | Yes |
| 81 | Arfgef2 | 11566 | -0.437 | -0.1691 | Yes |
| 82 | Tom1l1 | 11624 | -0.448 | -0.1531 | Yes |
| 83 | Snap23 | 11873 | -0.501 | -0.1502 | Yes |
| 84 | Scamp3 | 11964 | -0.532 | -0.1330 | Yes |
| 85 | Sspn | 12100 | -0.585 | -0.1169 | Yes |
| 86 | Krt18 | 12118 | -0.596 | -0.0908 | Yes |
| 87 | Bnip3 | 12239 | -0.701 | -0.0683 | Yes |
| 88 | Clta | 12282 | -0.753 | -0.0369 | Yes |
| 89 | Cd63 | 12345 | -0.941 | 0.0015 | Yes |

