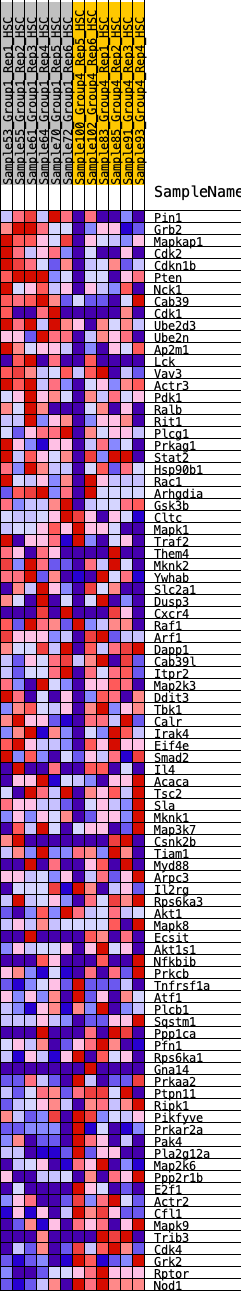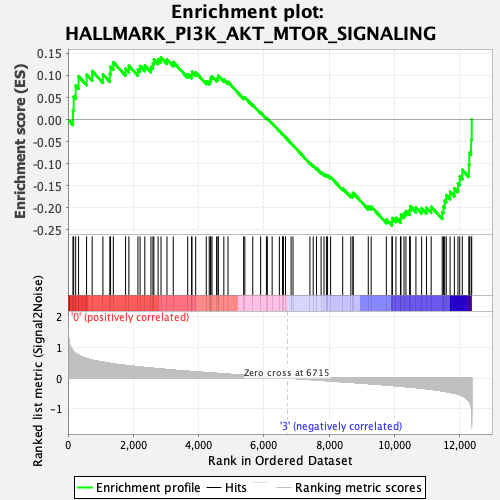
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.24077664 |
| Normalized Enrichment Score (NES) | -1.0287923 |
| Nominal p-value | 0.401222 |
| FDR q-value | 0.9661139 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pin1 | 151 | 0.899 | 0.0213 | No |
| 2 | Grb2 | 174 | 0.865 | 0.0519 | No |
| 3 | Mapkap1 | 235 | 0.801 | 0.0769 | No |
| 4 | Cdk2 | 323 | 0.751 | 0.0980 | No |
| 5 | Cdkn1b | 569 | 0.638 | 0.1019 | No |
| 6 | Pten | 739 | 0.587 | 0.1101 | No |
| 7 | Nck1 | 1069 | 0.519 | 0.1027 | No |
| 8 | Cab39 | 1281 | 0.480 | 0.1035 | No |
| 9 | Cdk1 | 1303 | 0.476 | 0.1196 | No |
| 10 | Ube2d3 | 1388 | 0.462 | 0.1300 | No |
| 11 | Ube2n | 1762 | 0.413 | 0.1151 | No |
| 12 | Ap2m1 | 1864 | 0.401 | 0.1218 | No |
| 13 | Lck | 2141 | 0.370 | 0.1132 | No |
| 14 | Vav3 | 2207 | 0.362 | 0.1214 | No |
| 15 | Actr3 | 2352 | 0.346 | 0.1226 | No |
| 16 | Pdk1 | 2540 | 0.326 | 0.1196 | No |
| 17 | Ralb | 2600 | 0.320 | 0.1268 | No |
| 18 | Rit1 | 2629 | 0.317 | 0.1364 | No |
| 19 | Plcg1 | 2759 | 0.302 | 0.1372 | No |
| 20 | Prkag1 | 2848 | 0.294 | 0.1410 | No |
| 21 | Stat2 | 3030 | 0.276 | 0.1365 | No |
| 22 | Hsp90b1 | 3222 | 0.257 | 0.1306 | No |
| 23 | Rac1 | 3665 | 0.223 | 0.1029 | No |
| 24 | Arhgdia | 3783 | 0.213 | 0.1013 | No |
| 25 | Gsk3b | 3796 | 0.212 | 0.1083 | No |
| 26 | Cltc | 3909 | 0.204 | 0.1068 | No |
| 27 | Mapk1 | 4233 | 0.177 | 0.0871 | No |
| 28 | Traf2 | 4324 | 0.170 | 0.0861 | No |
| 29 | Them4 | 4357 | 0.167 | 0.0898 | No |
| 30 | Mknk2 | 4373 | 0.166 | 0.0948 | No |
| 31 | Ywhab | 4409 | 0.163 | 0.0980 | No |
| 32 | Slc2a1 | 4549 | 0.154 | 0.0924 | No |
| 33 | Dusp3 | 4593 | 0.151 | 0.0946 | No |
| 34 | Cxcr4 | 4601 | 0.150 | 0.0996 | No |
| 35 | Raf1 | 4771 | 0.137 | 0.0910 | No |
| 36 | Arf1 | 4899 | 0.126 | 0.0853 | No |
| 37 | Dapp1 | 5377 | 0.092 | 0.0499 | No |
| 38 | Cab39l | 5414 | 0.089 | 0.0503 | No |
| 39 | Itpr2 | 5656 | 0.071 | 0.0333 | No |
| 40 | Map2k3 | 5898 | 0.055 | 0.0157 | No |
| 41 | Ddit3 | 6075 | 0.041 | 0.0029 | No |
| 42 | Tbk1 | 6102 | 0.039 | 0.0023 | No |
| 43 | Calr | 6250 | 0.031 | -0.0086 | No |
| 44 | Irak4 | 6471 | 0.015 | -0.0259 | No |
| 45 | Eif4e | 6566 | 0.009 | -0.0333 | No |
| 46 | Smad2 | 6596 | 0.007 | -0.0354 | No |
| 47 | Il4 | 6659 | 0.003 | -0.0403 | No |
| 48 | Acaca | 6830 | -0.005 | -0.0540 | No |
| 49 | Tsc2 | 6889 | -0.009 | -0.0584 | No |
| 50 | Sla | 7407 | -0.045 | -0.0988 | No |
| 51 | Mknk1 | 7507 | -0.052 | -0.1049 | No |
| 52 | Map3k7 | 7607 | -0.059 | -0.1108 | No |
| 53 | Csnk2b | 7749 | -0.070 | -0.1197 | No |
| 54 | Tiam1 | 7837 | -0.077 | -0.1239 | No |
| 55 | Myd88 | 7912 | -0.081 | -0.1269 | No |
| 56 | Arpc3 | 7947 | -0.084 | -0.1265 | No |
| 57 | Il2rg | 8040 | -0.092 | -0.1306 | No |
| 58 | Rps6ka3 | 8410 | -0.119 | -0.1562 | No |
| 59 | Akt1 | 8660 | -0.136 | -0.1714 | No |
| 60 | Mapk8 | 8723 | -0.141 | -0.1711 | No |
| 61 | Ecsit | 8731 | -0.141 | -0.1664 | No |
| 62 | Akt1s1 | 9193 | -0.178 | -0.1973 | No |
| 63 | Nfkbib | 9282 | -0.186 | -0.1975 | No |
| 64 | Prkcb | 9746 | -0.222 | -0.2269 | No |
| 65 | Tnfrsf1a | 9917 | -0.238 | -0.2319 | Yes |
| 66 | Atf1 | 9931 | -0.240 | -0.2240 | Yes |
| 67 | Plcb1 | 10040 | -0.251 | -0.2234 | Yes |
| 68 | Sqstm1 | 10179 | -0.262 | -0.2248 | Yes |
| 69 | Ppp1ca | 10194 | -0.264 | -0.2161 | Yes |
| 70 | Pfn1 | 10287 | -0.273 | -0.2134 | Yes |
| 71 | Rps6ka1 | 10347 | -0.279 | -0.2078 | Yes |
| 72 | Gna14 | 10458 | -0.288 | -0.2060 | Yes |
| 73 | Prkaa2 | 10481 | -0.290 | -0.1969 | Yes |
| 74 | Ptpn11 | 10652 | -0.309 | -0.1992 | Yes |
| 75 | Ripk1 | 10827 | -0.328 | -0.2011 | Yes |
| 76 | Pikfyve | 10975 | -0.346 | -0.2001 | Yes |
| 77 | Prkar2a | 11119 | -0.364 | -0.1982 | Yes |
| 78 | Pak4 | 11457 | -0.416 | -0.2101 | Yes |
| 79 | Pla2g12a | 11497 | -0.423 | -0.1974 | Yes |
| 80 | Map2k6 | 11533 | -0.429 | -0.1842 | Yes |
| 81 | Ppp2r1b | 11580 | -0.440 | -0.1715 | Yes |
| 82 | E2f1 | 11698 | -0.463 | -0.1637 | Yes |
| 83 | Actr2 | 11829 | -0.492 | -0.1559 | Yes |
| 84 | Cfl1 | 11939 | -0.522 | -0.1452 | Yes |
| 85 | Mapk9 | 11993 | -0.541 | -0.1293 | Yes |
| 86 | Trib3 | 12072 | -0.576 | -0.1141 | Yes |
| 87 | Cdk4 | 12274 | -0.745 | -0.1026 | Yes |
| 88 | Grk2 | 12288 | -0.758 | -0.0753 | Yes |
| 89 | Rptor | 12342 | -0.901 | -0.0459 | Yes |
| 90 | Nod1 | 12360 | -1.271 | 0.0002 | Yes |