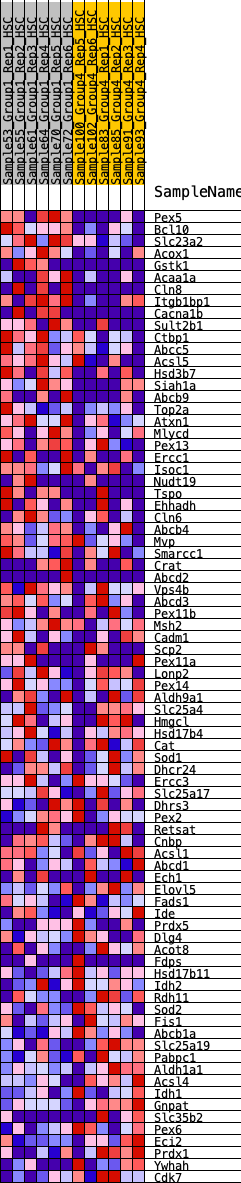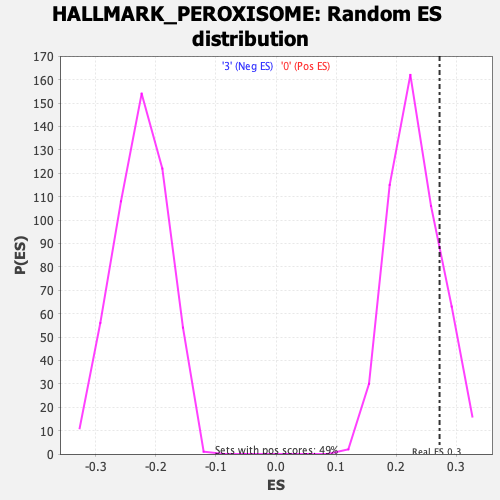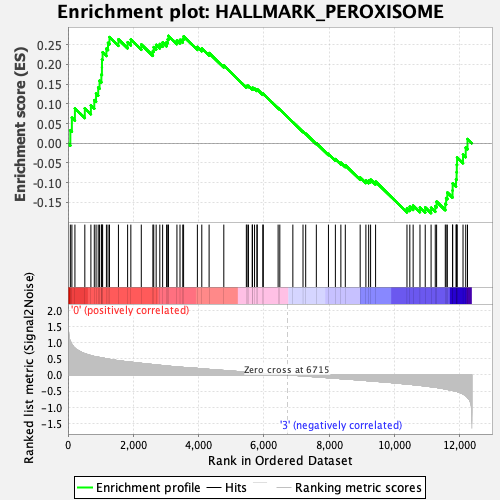
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.27232707 |
| Normalized Enrichment Score (NES) | 1.1787311 |
| Nominal p-value | 0.1680162 |
| FDR q-value | 0.5448459 |
| FWER p-Value | 0.944 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 72 | 1.032 | 0.0331 | Yes |
| 2 | Bcl10 | 119 | 0.942 | 0.0649 | Yes |
| 3 | Slc23a2 | 212 | 0.825 | 0.0886 | Yes |
| 4 | Acox1 | 515 | 0.654 | 0.0887 | Yes |
| 5 | Gstk1 | 701 | 0.598 | 0.0962 | Yes |
| 6 | Acaa1a | 805 | 0.570 | 0.1093 | Yes |
| 7 | Cln8 | 859 | 0.560 | 0.1262 | Yes |
| 8 | Itgb1bp1 | 925 | 0.548 | 0.1416 | Yes |
| 9 | Cacna1b | 966 | 0.540 | 0.1587 | Yes |
| 10 | Sult2b1 | 1022 | 0.530 | 0.1742 | Yes |
| 11 | Ctbp1 | 1039 | 0.525 | 0.1927 | Yes |
| 12 | Abcc5 | 1041 | 0.525 | 0.2124 | Yes |
| 13 | Acsl5 | 1060 | 0.521 | 0.2306 | Yes |
| 14 | Hsd3b7 | 1176 | 0.499 | 0.2401 | Yes |
| 15 | Siah1a | 1225 | 0.488 | 0.2546 | Yes |
| 16 | Abcb9 | 1270 | 0.481 | 0.2692 | Yes |
| 17 | Top2a | 1544 | 0.438 | 0.2635 | Yes |
| 18 | Atxn1 | 1825 | 0.405 | 0.2560 | Yes |
| 19 | Mlycd | 1924 | 0.396 | 0.2630 | Yes |
| 20 | Pex13 | 2244 | 0.358 | 0.2505 | Yes |
| 21 | Ercc1 | 2599 | 0.320 | 0.2338 | Yes |
| 22 | Isoc1 | 2625 | 0.317 | 0.2437 | Yes |
| 23 | Nudt19 | 2699 | 0.310 | 0.2495 | Yes |
| 24 | Tspo | 2811 | 0.297 | 0.2517 | Yes |
| 25 | Ehhadh | 2897 | 0.290 | 0.2557 | Yes |
| 26 | Cln6 | 3018 | 0.277 | 0.2563 | Yes |
| 27 | Abcb4 | 3055 | 0.273 | 0.2637 | Yes |
| 28 | Mvp | 3076 | 0.271 | 0.2723 | Yes |
| 29 | Smarcc1 | 3335 | 0.248 | 0.2607 | No |
| 30 | Crat | 3429 | 0.241 | 0.2622 | No |
| 31 | Abcd2 | 3514 | 0.233 | 0.2642 | No |
| 32 | Vps4b | 3539 | 0.231 | 0.2709 | No |
| 33 | Abcd3 | 3961 | 0.201 | 0.2442 | No |
| 34 | Pex11b | 4096 | 0.190 | 0.2405 | No |
| 35 | Msh2 | 4320 | 0.170 | 0.2287 | No |
| 36 | Cadm1 | 4770 | 0.137 | 0.1974 | No |
| 37 | Scp2 | 5460 | 0.085 | 0.1445 | No |
| 38 | Pex11a | 5483 | 0.083 | 0.1458 | No |
| 39 | Lonp2 | 5525 | 0.080 | 0.1455 | No |
| 40 | Pex14 | 5640 | 0.072 | 0.1390 | No |
| 41 | Aldh9a1 | 5648 | 0.072 | 0.1411 | No |
| 42 | Slc25a4 | 5711 | 0.068 | 0.1386 | No |
| 43 | Hmgcl | 5785 | 0.063 | 0.1350 | No |
| 44 | Hsd17b4 | 5793 | 0.062 | 0.1368 | No |
| 45 | Cat | 5964 | 0.050 | 0.1248 | No |
| 46 | Sod1 | 5979 | 0.048 | 0.1255 | No |
| 47 | Dhcr24 | 6433 | 0.018 | 0.0893 | No |
| 48 | Ercc3 | 6485 | 0.015 | 0.0857 | No |
| 49 | Slc25a17 | 6884 | -0.008 | 0.0536 | No |
| 50 | Dhrs3 | 7195 | -0.030 | 0.0295 | No |
| 51 | Pex2 | 7277 | -0.036 | 0.0243 | No |
| 52 | Retsat | 7603 | -0.059 | 0.0001 | No |
| 53 | Cnbp | 7974 | -0.086 | -0.0268 | No |
| 54 | Acsl1 | 8186 | -0.102 | -0.0401 | No |
| 55 | Abcd1 | 8354 | -0.115 | -0.0494 | No |
| 56 | Ech1 | 8490 | -0.125 | -0.0557 | No |
| 57 | Elovl5 | 8945 | -0.158 | -0.0867 | No |
| 58 | Fads1 | 9120 | -0.172 | -0.0943 | No |
| 59 | Ide | 9205 | -0.180 | -0.0944 | No |
| 60 | Prdx5 | 9264 | -0.185 | -0.0921 | No |
| 61 | Dlg4 | 9417 | -0.196 | -0.0971 | No |
| 62 | Acot8 | 10378 | -0.281 | -0.1647 | No |
| 63 | Fdps | 10465 | -0.288 | -0.1608 | No |
| 64 | Hsd17b11 | 10567 | -0.299 | -0.1577 | No |
| 65 | Idh2 | 10776 | -0.323 | -0.1624 | No |
| 66 | Rdh11 | 10937 | -0.341 | -0.1626 | No |
| 67 | Sod2 | 11116 | -0.363 | -0.1634 | No |
| 68 | Fis1 | 11244 | -0.381 | -0.1594 | No |
| 69 | Abcb1a | 11285 | -0.387 | -0.1480 | No |
| 70 | Slc25a19 | 11551 | -0.433 | -0.1532 | No |
| 71 | Pabpc1 | 11577 | -0.439 | -0.1387 | No |
| 72 | Aldh1a1 | 11614 | -0.446 | -0.1248 | No |
| 73 | Acsl4 | 11773 | -0.478 | -0.1196 | No |
| 74 | Idh1 | 11777 | -0.479 | -0.1018 | No |
| 75 | Gnpat | 11880 | -0.503 | -0.0911 | No |
| 76 | Slc35b2 | 11898 | -0.507 | -0.0733 | No |
| 77 | Pex6 | 11904 | -0.508 | -0.0546 | No |
| 78 | Eci2 | 11912 | -0.512 | -0.0358 | No |
| 79 | Prdx1 | 12094 | -0.584 | -0.0285 | No |
| 80 | Ywhah | 12177 | -0.638 | -0.0111 | No |
| 81 | Cdk7 | 12231 | -0.693 | 0.0107 | No |