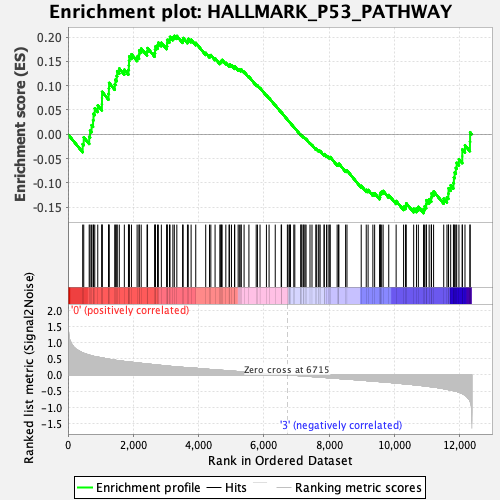
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_P53_PATHWAY |
| Enrichment Score (ES) | 0.2023727 |
| Normalized Enrichment Score (NES) | 0.89191884 |
| Nominal p-value | 0.66191447 |
| FDR q-value | 0.92923415 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plk2 | 450 | 0.681 | -0.0201 | Yes |
| 2 | Baiap2 | 482 | 0.668 | -0.0063 | Yes |
| 3 | Blcap | 649 | 0.612 | -0.0048 | Yes |
| 4 | Ier5 | 676 | 0.605 | 0.0079 | Yes |
| 5 | Csrnp2 | 722 | 0.591 | 0.0187 | Yes |
| 6 | St14 | 767 | 0.577 | 0.0293 | Yes |
| 7 | Tgfb1 | 781 | 0.575 | 0.0423 | Yes |
| 8 | Pom121 | 819 | 0.567 | 0.0532 | Yes |
| 9 | Tcn2 | 911 | 0.551 | 0.0593 | Yes |
| 10 | Ccng1 | 1038 | 0.526 | 0.0619 | Yes |
| 11 | Abcc5 | 1041 | 0.525 | 0.0746 | Yes |
| 12 | Cyfip2 | 1043 | 0.525 | 0.0874 | Yes |
| 13 | Hdac3 | 1242 | 0.485 | 0.0831 | Yes |
| 14 | Abhd4 | 1252 | 0.484 | 0.0943 | Yes |
| 15 | Ctsd | 1258 | 0.483 | 0.1057 | Yes |
| 16 | Ppp1r15a | 1432 | 0.454 | 0.1027 | Yes |
| 17 | Pcna | 1450 | 0.452 | 0.1124 | Yes |
| 18 | Gm2a | 1492 | 0.446 | 0.1200 | Yes |
| 19 | Dgka | 1506 | 0.444 | 0.1299 | Yes |
| 20 | Foxo3 | 1570 | 0.435 | 0.1354 | Yes |
| 21 | Pitpnc1 | 1725 | 0.416 | 0.1330 | Yes |
| 22 | Dram1 | 1851 | 0.402 | 0.1326 | Yes |
| 23 | Tprkb | 1863 | 0.401 | 0.1416 | Yes |
| 24 | Ddb2 | 1868 | 0.401 | 0.1511 | Yes |
| 25 | Prmt2 | 1876 | 0.400 | 0.1603 | Yes |
| 26 | Sec61a1 | 1943 | 0.393 | 0.1646 | Yes |
| 27 | Fam162a | 2116 | 0.375 | 0.1597 | Yes |
| 28 | Slc35d1 | 2175 | 0.366 | 0.1639 | Yes |
| 29 | Zmat3 | 2180 | 0.365 | 0.1726 | Yes |
| 30 | Rrp8 | 2241 | 0.358 | 0.1764 | Yes |
| 31 | Tob1 | 2420 | 0.338 | 0.1702 | Yes |
| 32 | Stom | 2435 | 0.337 | 0.1773 | Yes |
| 33 | Pvt1 | 2653 | 0.315 | 0.1672 | Yes |
| 34 | Vwa5a | 2665 | 0.313 | 0.1740 | Yes |
| 35 | S100a10 | 2678 | 0.312 | 0.1807 | Yes |
| 36 | Osgin1 | 2741 | 0.304 | 0.1831 | Yes |
| 37 | Ldhb | 2766 | 0.301 | 0.1885 | Yes |
| 38 | App | 2858 | 0.293 | 0.1883 | Yes |
| 39 | Jag2 | 3020 | 0.277 | 0.1819 | Yes |
| 40 | Ndrg1 | 3035 | 0.275 | 0.1875 | Yes |
| 41 | Wrap73 | 3037 | 0.275 | 0.1942 | Yes |
| 42 | Rxra | 3107 | 0.268 | 0.1951 | Yes |
| 43 | Polh | 3123 | 0.266 | 0.2004 | Yes |
| 44 | Ercc5 | 3211 | 0.258 | 0.1996 | Yes |
| 45 | Pidd1 | 3255 | 0.255 | 0.2024 | Yes |
| 46 | Ctsf | 3332 | 0.249 | 0.2023 | No |
| 47 | Iscu | 3508 | 0.233 | 0.1936 | No |
| 48 | Ip6k2 | 3525 | 0.232 | 0.1980 | No |
| 49 | Ddit4 | 3657 | 0.223 | 0.1928 | No |
| 50 | Eps8l2 | 3681 | 0.222 | 0.1963 | No |
| 51 | Cgrrf1 | 3769 | 0.214 | 0.1945 | No |
| 52 | Ier3 | 3911 | 0.204 | 0.1879 | No |
| 53 | Mapkapk3 | 4216 | 0.179 | 0.1674 | No |
| 54 | Tpd52l1 | 4333 | 0.169 | 0.1621 | No |
| 55 | Mknk2 | 4373 | 0.166 | 0.1629 | No |
| 56 | Rchy1 | 4503 | 0.158 | 0.1562 | No |
| 57 | Ccnd2 | 4649 | 0.146 | 0.1480 | No |
| 58 | Plxnb2 | 4674 | 0.143 | 0.1495 | No |
| 59 | Fbxw7 | 4700 | 0.142 | 0.1509 | No |
| 60 | Dcxr | 4718 | 0.140 | 0.1530 | No |
| 61 | Sesn1 | 4832 | 0.132 | 0.1470 | No |
| 62 | Upp1 | 4930 | 0.124 | 0.1421 | No |
| 63 | Txnip | 4947 | 0.123 | 0.1438 | No |
| 64 | Notch1 | 5006 | 0.118 | 0.1420 | No |
| 65 | Vdr | 5099 | 0.112 | 0.1372 | No |
| 66 | Wwp1 | 5104 | 0.112 | 0.1396 | No |
| 67 | Acvr1b | 5209 | 0.103 | 0.1336 | No |
| 68 | Ada | 5245 | 0.100 | 0.1332 | No |
| 69 | Zbtb16 | 5293 | 0.097 | 0.1318 | No |
| 70 | Btg1 | 5303 | 0.097 | 0.1334 | No |
| 71 | Mdm2 | 5391 | 0.091 | 0.1285 | No |
| 72 | Jun | 5539 | 0.079 | 0.1184 | No |
| 73 | Bak1 | 5762 | 0.065 | 0.1018 | No |
| 74 | Rhbdf2 | 5800 | 0.062 | 0.1003 | No |
| 75 | Ccnk | 5879 | 0.056 | 0.0953 | No |
| 76 | Ddit3 | 6075 | 0.041 | 0.0803 | No |
| 77 | Ralgds | 6156 | 0.036 | 0.0747 | No |
| 78 | Sertad3 | 6345 | 0.024 | 0.0598 | No |
| 79 | Rab40c | 6527 | 0.012 | 0.0453 | No |
| 80 | Irag2 | 6538 | 0.010 | 0.0447 | No |
| 81 | Klf4 | 6716 | 0.000 | 0.0303 | No |
| 82 | Abat | 6738 | 0.000 | 0.0285 | No |
| 83 | Ninj1 | 6787 | -0.002 | 0.0247 | No |
| 84 | Fuca1 | 6793 | -0.003 | 0.0243 | No |
| 85 | Ak1 | 6812 | -0.004 | 0.0229 | No |
| 86 | Rad9a | 6913 | -0.010 | 0.0150 | No |
| 87 | Zfp36l1 | 6944 | -0.012 | 0.0128 | No |
| 88 | H2ac25 | 7129 | -0.025 | -0.0016 | No |
| 89 | H2aj | 7132 | -0.025 | -0.0012 | No |
| 90 | Tm4sf1 | 7161 | -0.027 | -0.0028 | No |
| 91 | Apaf1 | 7204 | -0.031 | -0.0055 | No |
| 92 | Steap3 | 7224 | -0.032 | -0.0062 | No |
| 93 | Sat1 | 7257 | -0.034 | -0.0080 | No |
| 94 | Coq8a | 7288 | -0.036 | -0.0096 | No |
| 95 | Bax | 7409 | -0.045 | -0.0183 | No |
| 96 | Ptpre | 7470 | -0.050 | -0.0220 | No |
| 97 | Rnf19b | 7588 | -0.058 | -0.0301 | No |
| 98 | Retsat | 7603 | -0.059 | -0.0299 | No |
| 99 | Irak1 | 7661 | -0.064 | -0.0330 | No |
| 100 | Trp53 | 7685 | -0.065 | -0.0332 | No |
| 101 | Ephx1 | 7720 | -0.068 | -0.0344 | No |
| 102 | Slc3a2 | 7840 | -0.077 | -0.0422 | No |
| 103 | H1f2 | 7845 | -0.077 | -0.0406 | No |
| 104 | Fos | 7915 | -0.082 | -0.0443 | No |
| 105 | Def6 | 7964 | -0.085 | -0.0461 | No |
| 106 | Hbegf | 8013 | -0.090 | -0.0479 | No |
| 107 | Hint1 | 8027 | -0.090 | -0.0467 | No |
| 108 | Ccp110 | 8240 | -0.106 | -0.0615 | No |
| 109 | Trafd1 | 8279 | -0.109 | -0.0619 | No |
| 110 | Ei24 | 8294 | -0.110 | -0.0604 | No |
| 111 | Ppm1d | 8499 | -0.126 | -0.0740 | No |
| 112 | Mxd4 | 8541 | -0.129 | -0.0742 | No |
| 113 | Cd82 | 8977 | -0.160 | -0.1059 | No |
| 114 | Prkab1 | 9131 | -0.173 | -0.1141 | No |
| 115 | Rb1 | 9188 | -0.178 | -0.1144 | No |
| 116 | Tsc22d1 | 9334 | -0.189 | -0.1216 | No |
| 117 | Tspyl2 | 9388 | -0.194 | -0.1212 | No |
| 118 | Tgfa | 9540 | -0.206 | -0.1285 | No |
| 119 | F2r | 9554 | -0.207 | -0.1245 | No |
| 120 | Cdkn2aip | 9565 | -0.208 | -0.1202 | No |
| 121 | Fdxr | 9603 | -0.211 | -0.1180 | No |
| 122 | Hspa4l | 9648 | -0.213 | -0.1164 | No |
| 123 | Tm7sf3 | 9820 | -0.229 | -0.1248 | No |
| 124 | Rps12 | 10046 | -0.251 | -0.1370 | No |
| 125 | Socs1 | 10274 | -0.271 | -0.1490 | No |
| 126 | Hexim1 | 10332 | -0.277 | -0.1468 | No |
| 127 | Aen | 10356 | -0.280 | -0.1419 | No |
| 128 | Vamp8 | 10583 | -0.301 | -0.1530 | No |
| 129 | Procr | 10669 | -0.311 | -0.1523 | No |
| 130 | Casp1 | 10726 | -0.317 | -0.1491 | No |
| 131 | Fgf13 | 10885 | -0.334 | -0.1538 | No |
| 132 | Slc19a2 | 10918 | -0.339 | -0.1481 | No |
| 133 | Kif13b | 10966 | -0.345 | -0.1435 | No |
| 134 | Rap2b | 10972 | -0.346 | -0.1354 | No |
| 135 | Mxd1 | 11057 | -0.358 | -0.1335 | No |
| 136 | Elp1 | 11121 | -0.364 | -0.1298 | No |
| 137 | Nol8 | 11129 | -0.365 | -0.1214 | No |
| 138 | Fas | 11193 | -0.374 | -0.1174 | No |
| 139 | Klk8 | 11503 | -0.424 | -0.1322 | No |
| 140 | Cd81 | 11602 | -0.444 | -0.1294 | No |
| 141 | Rpl36 | 11645 | -0.453 | -0.1217 | No |
| 142 | Xpc | 11651 | -0.454 | -0.1110 | No |
| 143 | Rack1 | 11716 | -0.467 | -0.1047 | No |
| 144 | Tap1 | 11800 | -0.485 | -0.0996 | No |
| 145 | Dnttip2 | 11815 | -0.489 | -0.0888 | No |
| 146 | Rps27l | 11838 | -0.493 | -0.0784 | No |
| 147 | Ptpn14 | 11874 | -0.502 | -0.0690 | No |
| 148 | Rpl18 | 11900 | -0.508 | -0.0586 | No |
| 149 | Ankra2 | 11969 | -0.533 | -0.0511 | No |
| 150 | Ccnd3 | 12071 | -0.575 | -0.0452 | No |
| 151 | Trib3 | 12072 | -0.576 | -0.0311 | No |
| 152 | Sp1 | 12154 | -0.621 | -0.0224 | No |
| 153 | Nupr1 | 12306 | -0.798 | -0.0152 | No |
| 154 | Pmm1 | 12311 | -0.807 | 0.0043 | No |

