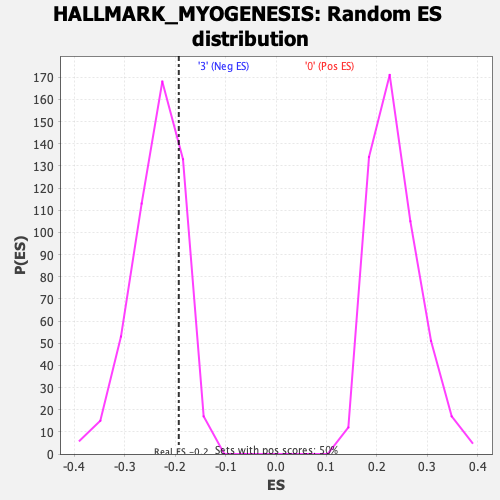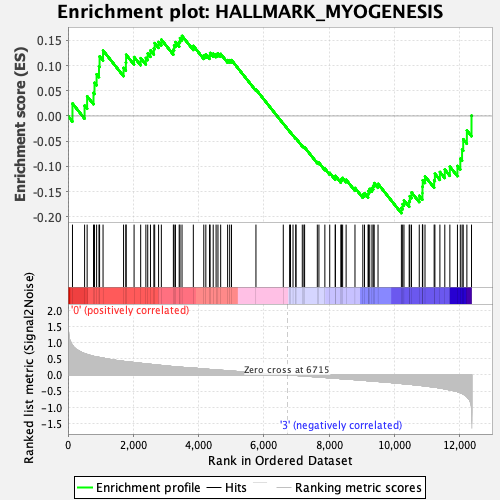
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.19296035 |
| Normalized Enrichment Score (NES) | -0.82058656 |
| Nominal p-value | 0.7980198 |
| FDR q-value | 0.8710379 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myh9 | 137 | 0.913 | 0.0248 | No |
| 2 | Mef2d | 508 | 0.657 | 0.0206 | No |
| 3 | Eno3 | 586 | 0.630 | 0.0391 | No |
| 4 | Tgfb1 | 781 | 0.575 | 0.0460 | No |
| 5 | Mapk12 | 812 | 0.568 | 0.0659 | No |
| 6 | Aebp1 | 876 | 0.558 | 0.0828 | No |
| 7 | Sh2b1 | 949 | 0.543 | 0.0983 | No |
| 8 | Gadd45b | 967 | 0.540 | 0.1182 | No |
| 9 | Sirt2 | 1073 | 0.519 | 0.1301 | No |
| 10 | Ocel1 | 1701 | 0.419 | 0.0955 | No |
| 11 | Gsn | 1772 | 0.411 | 0.1061 | No |
| 12 | Ptp4a3 | 1779 | 0.411 | 0.1218 | No |
| 13 | Akt2 | 2023 | 0.383 | 0.1170 | No |
| 14 | Sptan1 | 2225 | 0.360 | 0.1148 | No |
| 15 | Pdlim7 | 2384 | 0.341 | 0.1154 | No |
| 16 | Gaa | 2441 | 0.336 | 0.1241 | No |
| 17 | Eif4a2 | 2525 | 0.327 | 0.1302 | No |
| 18 | Rit1 | 2629 | 0.317 | 0.1343 | No |
| 19 | Flii | 2654 | 0.314 | 0.1448 | No |
| 20 | Agl | 2773 | 0.300 | 0.1470 | No |
| 21 | App | 2858 | 0.293 | 0.1517 | No |
| 22 | Lpin1 | 3226 | 0.257 | 0.1319 | No |
| 23 | Itgb1 | 3248 | 0.255 | 0.1403 | No |
| 24 | Tpm3 | 3290 | 0.252 | 0.1469 | No |
| 25 | Ncam1 | 3404 | 0.244 | 0.1473 | No |
| 26 | Crat | 3429 | 0.241 | 0.1548 | No |
| 27 | Tpm2 | 3491 | 0.234 | 0.1591 | No |
| 28 | Speg | 3837 | 0.209 | 0.1392 | No |
| 29 | Pygm | 4157 | 0.184 | 0.1205 | No |
| 30 | Hdac5 | 4220 | 0.179 | 0.1225 | No |
| 31 | Tpd52l1 | 4333 | 0.169 | 0.1200 | No |
| 32 | Lsp1 | 4351 | 0.167 | 0.1252 | No |
| 33 | Adcy9 | 4446 | 0.161 | 0.1239 | No |
| 34 | Sorbs3 | 4534 | 0.155 | 0.1229 | No |
| 35 | Ryr1 | 4591 | 0.151 | 0.1243 | No |
| 36 | Plxnb2 | 4674 | 0.143 | 0.1233 | No |
| 37 | Pick1 | 4887 | 0.127 | 0.1110 | No |
| 38 | Chrnb1 | 4950 | 0.123 | 0.1108 | No |
| 39 | Notch1 | 5006 | 0.118 | 0.1110 | No |
| 40 | Foxo4 | 5754 | 0.065 | 0.0527 | No |
| 41 | Pcx | 6591 | 0.007 | -0.0151 | No |
| 42 | Pfkm | 6785 | -0.002 | -0.0308 | No |
| 43 | Ak1 | 6812 | -0.004 | -0.0327 | No |
| 44 | Tsc2 | 6889 | -0.009 | -0.0386 | No |
| 45 | Igf1 | 6973 | -0.014 | -0.0448 | No |
| 46 | Ache | 6981 | -0.015 | -0.0448 | No |
| 47 | Erbb3 | 7173 | -0.028 | -0.0592 | No |
| 48 | Dtna | 7223 | -0.032 | -0.0620 | No |
| 49 | Psen2 | 7244 | -0.034 | -0.0623 | No |
| 50 | Gabarapl2 | 7633 | -0.061 | -0.0915 | No |
| 51 | Gpx3 | 7682 | -0.065 | -0.0928 | No |
| 52 | Dmd | 7864 | -0.078 | -0.1045 | No |
| 53 | Hbegf | 8013 | -0.090 | -0.1130 | No |
| 54 | Bin1 | 8182 | -0.102 | -0.1227 | No |
| 55 | Acsl1 | 8186 | -0.102 | -0.1189 | No |
| 56 | Dmpk | 8353 | -0.115 | -0.1279 | No |
| 57 | Ablim1 | 8374 | -0.116 | -0.1250 | No |
| 58 | Mylk | 8408 | -0.118 | -0.1230 | No |
| 59 | Fhl1 | 8515 | -0.127 | -0.1266 | No |
| 60 | Schip1 | 8786 | -0.145 | -0.1429 | No |
| 61 | Bhlhe40 | 9026 | -0.165 | -0.1559 | No |
| 62 | Camk2b | 9078 | -0.169 | -0.1534 | No |
| 63 | Rb1 | 9188 | -0.178 | -0.1552 | No |
| 64 | Myom1 | 9198 | -0.179 | -0.1489 | No |
| 65 | Svil | 9237 | -0.183 | -0.1448 | No |
| 66 | Gnao1 | 9306 | -0.188 | -0.1429 | No |
| 67 | Prnp | 9345 | -0.190 | -0.1385 | No |
| 68 | Mef2a | 9374 | -0.193 | -0.1332 | No |
| 69 | Wwtr1 | 9492 | -0.202 | -0.1348 | No |
| 70 | Clu | 10207 | -0.265 | -0.1825 | Yes |
| 71 | Cnn3 | 10246 | -0.268 | -0.1750 | Yes |
| 72 | Myo1c | 10286 | -0.273 | -0.1675 | Yes |
| 73 | Mapre3 | 10446 | -0.287 | -0.1691 | Yes |
| 74 | Fdps | 10465 | -0.288 | -0.1592 | Yes |
| 75 | Mras | 10517 | -0.293 | -0.1518 | Yes |
| 76 | Atp6ap1 | 10754 | -0.321 | -0.1584 | Yes |
| 77 | Smtn | 10851 | -0.331 | -0.1532 | Yes |
| 78 | Agrn | 10853 | -0.331 | -0.1402 | Yes |
| 79 | Col4a2 | 10861 | -0.332 | -0.1277 | Yes |
| 80 | Pde4dip | 10930 | -0.340 | -0.1198 | Yes |
| 81 | Ctf1 | 11210 | -0.376 | -0.1277 | Yes |
| 82 | Mef2c | 11233 | -0.379 | -0.1146 | Yes |
| 83 | Kcnh2 | 11386 | -0.402 | -0.1111 | Yes |
| 84 | Syngr2 | 11536 | -0.429 | -0.1064 | Yes |
| 85 | Bag1 | 11691 | -0.462 | -0.1007 | Yes |
| 86 | Pkia | 11923 | -0.515 | -0.0992 | Yes |
| 87 | Large1 | 12011 | -0.548 | -0.0847 | Yes |
| 88 | Ifrd1 | 12065 | -0.573 | -0.0664 | Yes |
| 89 | Sspn | 12100 | -0.585 | -0.0461 | Yes |
| 90 | Nav2 | 12212 | -0.670 | -0.0288 | Yes |
| 91 | Sorbs1 | 12354 | -1.039 | 0.0007 | Yes |