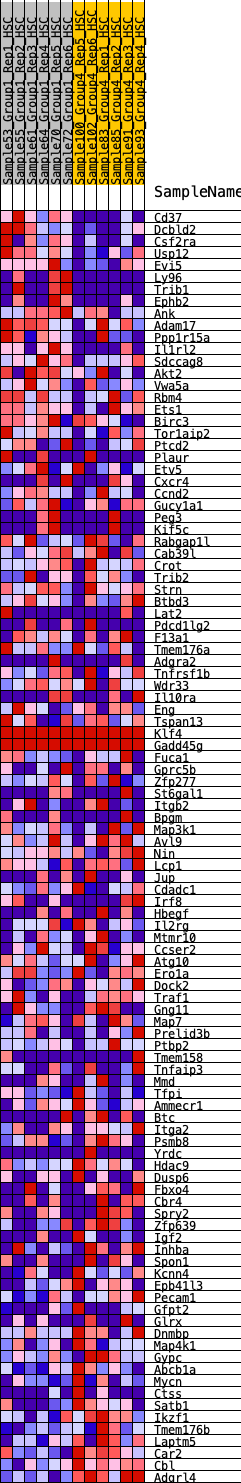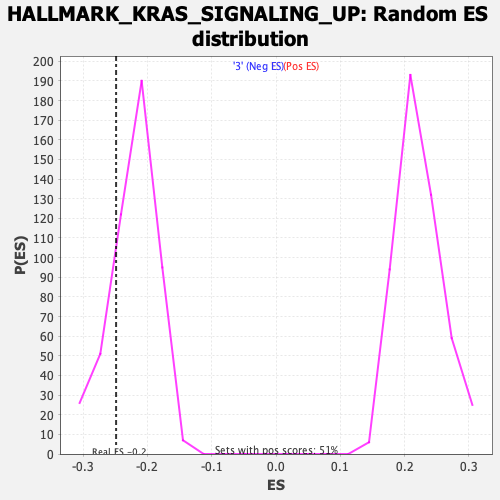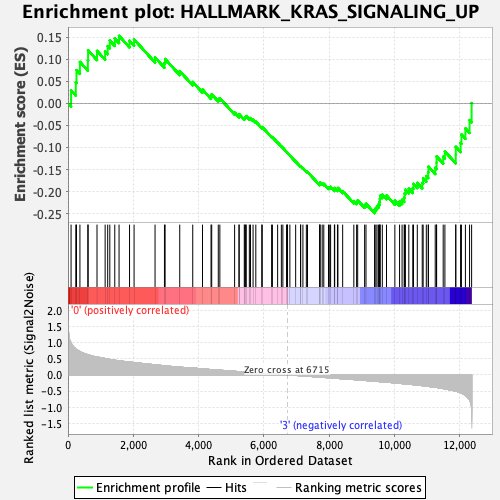
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.24873836 |
| Normalized Enrichment Score (NES) | -1.1238468 |
| Nominal p-value | 0.17922607 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cd37 | 94 | 0.989 | 0.0293 | No |
| 2 | Dcbld2 | 239 | 0.798 | 0.0475 | No |
| 3 | Csf2ra | 260 | 0.784 | 0.0752 | No |
| 4 | Usp12 | 365 | 0.723 | 0.0937 | No |
| 5 | Evi5 | 608 | 0.625 | 0.0974 | No |
| 6 | Ly96 | 616 | 0.623 | 0.1201 | No |
| 7 | Trib1 | 888 | 0.555 | 0.1188 | No |
| 8 | Ephb2 | 1137 | 0.507 | 0.1175 | No |
| 9 | Ank | 1215 | 0.491 | 0.1296 | No |
| 10 | Adam17 | 1280 | 0.480 | 0.1424 | No |
| 11 | Ppp1r15a | 1432 | 0.454 | 0.1471 | No |
| 12 | Il1rl2 | 1563 | 0.436 | 0.1528 | No |
| 13 | Sdccag8 | 1884 | 0.399 | 0.1416 | No |
| 14 | Akt2 | 2023 | 0.383 | 0.1447 | No |
| 15 | Vwa5a | 2665 | 0.313 | 0.1041 | No |
| 16 | Rbm4 | 2953 | 0.285 | 0.0914 | No |
| 17 | Ets1 | 2973 | 0.282 | 0.1004 | No |
| 18 | Birc3 | 3421 | 0.242 | 0.0729 | No |
| 19 | Tor1aip2 | 3819 | 0.210 | 0.0484 | No |
| 20 | Ptcd2 | 4118 | 0.188 | 0.0312 | No |
| 21 | Plaur | 4379 | 0.165 | 0.0161 | No |
| 22 | Etv5 | 4400 | 0.164 | 0.0206 | No |
| 23 | Cxcr4 | 4601 | 0.150 | 0.0099 | No |
| 24 | Ccnd2 | 4649 | 0.146 | 0.0115 | No |
| 25 | Gucy1a1 | 5101 | 0.112 | -0.0210 | No |
| 26 | Peg3 | 5237 | 0.101 | -0.0283 | No |
| 27 | Kif5c | 5238 | 0.101 | -0.0245 | No |
| 28 | Rabgap1l | 5397 | 0.090 | -0.0340 | No |
| 29 | Cab39l | 5414 | 0.089 | -0.0320 | No |
| 30 | Crot | 5437 | 0.088 | -0.0305 | No |
| 31 | Trib2 | 5457 | 0.085 | -0.0289 | No |
| 32 | Strn | 5552 | 0.078 | -0.0336 | No |
| 33 | Btbd3 | 5587 | 0.076 | -0.0336 | No |
| 34 | Lat2 | 5666 | 0.071 | -0.0373 | No |
| 35 | Pdcd1lg2 | 5750 | 0.066 | -0.0416 | No |
| 36 | F13a1 | 5932 | 0.052 | -0.0544 | No |
| 37 | Tmem176a | 5947 | 0.051 | -0.0537 | No |
| 38 | Adgra2 | 6234 | 0.031 | -0.0758 | No |
| 39 | Tnfrsf1b | 6258 | 0.030 | -0.0766 | No |
| 40 | Wdr33 | 6418 | 0.019 | -0.0888 | No |
| 41 | Il10ra | 6532 | 0.011 | -0.0976 | No |
| 42 | Eng | 6578 | 0.008 | -0.1010 | No |
| 43 | Tspan13 | 6691 | 0.001 | -0.1101 | No |
| 44 | Klf4 | 6716 | 0.000 | -0.1121 | No |
| 45 | Gadd45g | 6721 | 0.000 | -0.1124 | No |
| 46 | Fuca1 | 6793 | -0.003 | -0.1181 | No |
| 47 | Gprc5b | 6970 | -0.014 | -0.1319 | No |
| 48 | Zfp277 | 7120 | -0.025 | -0.1432 | No |
| 49 | St6gal1 | 7123 | -0.025 | -0.1424 | No |
| 50 | Itgb2 | 7192 | -0.030 | -0.1468 | No |
| 51 | Bpgm | 7297 | -0.037 | -0.1539 | No |
| 52 | Map3k1 | 7333 | -0.040 | -0.1553 | No |
| 53 | Avl9 | 7702 | -0.067 | -0.1828 | No |
| 54 | Nin | 7712 | -0.067 | -0.1810 | No |
| 55 | Lcp1 | 7718 | -0.068 | -0.1789 | No |
| 56 | Jup | 7782 | -0.072 | -0.1814 | No |
| 57 | Cdadc1 | 7831 | -0.076 | -0.1824 | No |
| 58 | Irf8 | 7975 | -0.086 | -0.1909 | No |
| 59 | Hbegf | 8013 | -0.090 | -0.1905 | No |
| 60 | Il2rg | 8040 | -0.092 | -0.1892 | No |
| 61 | Mtmr10 | 8156 | -0.100 | -0.1949 | No |
| 62 | Ccser2 | 8165 | -0.101 | -0.1917 | No |
| 63 | Atg10 | 8250 | -0.106 | -0.1946 | No |
| 64 | Ero1a | 8264 | -0.108 | -0.1916 | No |
| 65 | Dock2 | 8409 | -0.118 | -0.1989 | No |
| 66 | Traf1 | 8751 | -0.143 | -0.2214 | No |
| 67 | Gng11 | 8836 | -0.149 | -0.2227 | No |
| 68 | Map7 | 8869 | -0.151 | -0.2197 | No |
| 69 | Prelid3b | 9081 | -0.169 | -0.2306 | No |
| 70 | Ptbp2 | 9123 | -0.172 | -0.2274 | No |
| 71 | Tmem158 | 9385 | -0.194 | -0.2415 | Yes |
| 72 | Tnfaip3 | 9424 | -0.197 | -0.2372 | Yes |
| 73 | Mmd | 9461 | -0.199 | -0.2327 | Yes |
| 74 | Tfpi | 9508 | -0.203 | -0.2288 | Yes |
| 75 | Ammecr1 | 9536 | -0.206 | -0.2233 | Yes |
| 76 | Btc | 9549 | -0.207 | -0.2166 | Yes |
| 77 | Itga2 | 9557 | -0.207 | -0.2094 | Yes |
| 78 | Psmb8 | 9623 | -0.212 | -0.2068 | Yes |
| 79 | Yrdc | 9751 | -0.222 | -0.2088 | Yes |
| 80 | Hdac9 | 10007 | -0.248 | -0.2203 | Yes |
| 81 | Dusp6 | 10151 | -0.260 | -0.2223 | Yes |
| 82 | Fbxo4 | 10228 | -0.267 | -0.2185 | Yes |
| 83 | Cbr4 | 10292 | -0.273 | -0.2134 | Yes |
| 84 | Spry2 | 10305 | -0.274 | -0.2041 | Yes |
| 85 | Zfp639 | 10330 | -0.277 | -0.1957 | Yes |
| 86 | Igf2 | 10434 | -0.286 | -0.1934 | Yes |
| 87 | Inhba | 10556 | -0.298 | -0.1921 | Yes |
| 88 | Spon1 | 10577 | -0.300 | -0.1825 | Yes |
| 89 | Kcnn4 | 10694 | -0.314 | -0.1802 | Yes |
| 90 | Epb41l3 | 10845 | -0.330 | -0.1801 | Yes |
| 91 | Pecam1 | 10873 | -0.333 | -0.1698 | Yes |
| 92 | Gfpt2 | 10970 | -0.346 | -0.1647 | Yes |
| 93 | Glrx | 11028 | -0.354 | -0.1561 | Yes |
| 94 | Dnmbp | 11035 | -0.355 | -0.1433 | Yes |
| 95 | Map4k1 | 11243 | -0.381 | -0.1460 | Yes |
| 96 | Gypc | 11283 | -0.387 | -0.1347 | Yes |
| 97 | Abcb1a | 11285 | -0.387 | -0.1202 | Yes |
| 98 | Mycn | 11487 | -0.421 | -0.1209 | Yes |
| 99 | Ctss | 11539 | -0.430 | -0.1089 | Yes |
| 100 | Satb1 | 11871 | -0.500 | -0.1172 | Yes |
| 101 | Ikzf1 | 11872 | -0.501 | -0.0985 | Yes |
| 102 | Tmem176b | 12019 | -0.551 | -0.0898 | Yes |
| 103 | Laptm5 | 12045 | -0.566 | -0.0706 | Yes |
| 104 | Car2 | 12167 | -0.632 | -0.0568 | Yes |
| 105 | Cbl | 12294 | -0.775 | -0.0381 | Yes |
| 106 | Adgrl4 | 12357 | -1.166 | 0.0005 | Yes |