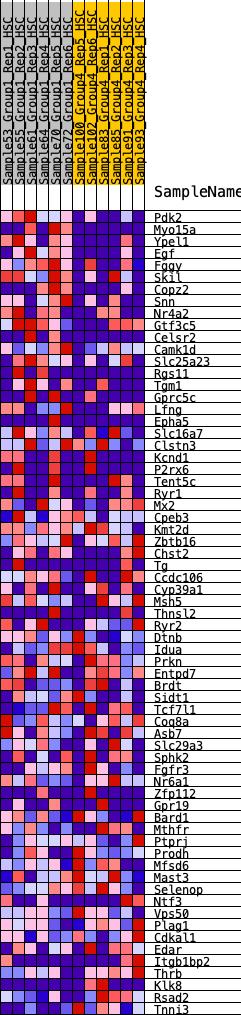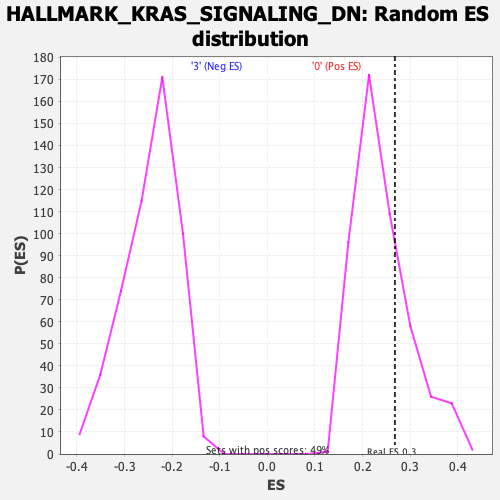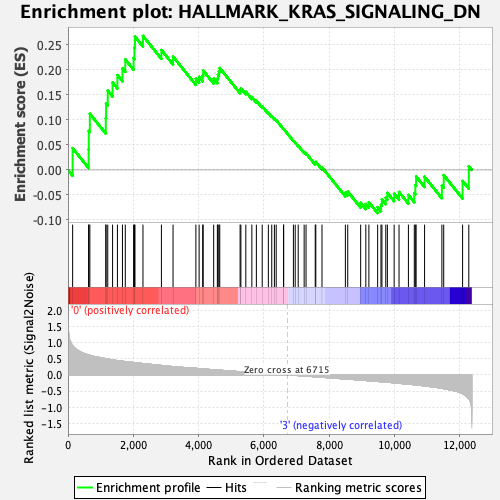
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2679549 |
| Normalized Enrichment Score (NES) | 1.1101573 |
| Nominal p-value | 0.26899385 |
| FDR q-value | 0.5470175 |
| FWER p-Value | 0.968 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pdk2 | 142 | 0.909 | 0.0435 | Yes |
| 2 | Myo15a | 632 | 0.616 | 0.0411 | Yes |
| 3 | Ypel1 | 636 | 0.616 | 0.0782 | Yes |
| 4 | Egf | 669 | 0.606 | 0.1123 | Yes |
| 5 | Fggy | 1157 | 0.503 | 0.1032 | Yes |
| 6 | Skil | 1166 | 0.501 | 0.1329 | Yes |
| 7 | Copz2 | 1217 | 0.490 | 0.1585 | Yes |
| 8 | Snn | 1365 | 0.465 | 0.1748 | Yes |
| 9 | Nr4a2 | 1513 | 0.444 | 0.1897 | Yes |
| 10 | Gtf3c5 | 1667 | 0.423 | 0.2029 | Yes |
| 11 | Celsr2 | 1754 | 0.413 | 0.2210 | Yes |
| 12 | Camk1d | 2008 | 0.384 | 0.2237 | Yes |
| 13 | Slc25a23 | 2035 | 0.381 | 0.2446 | Yes |
| 14 | Rgs11 | 2050 | 0.380 | 0.2665 | Yes |
| 15 | Tgm1 | 2295 | 0.352 | 0.2680 | Yes |
| 16 | Gprc5c | 2860 | 0.293 | 0.2398 | No |
| 17 | Lfng | 3216 | 0.258 | 0.2266 | No |
| 18 | Epha5 | 3915 | 0.204 | 0.1822 | No |
| 19 | Slc16a7 | 4014 | 0.197 | 0.1861 | No |
| 20 | Clstn3 | 4124 | 0.187 | 0.1886 | No |
| 21 | Kcnd1 | 4138 | 0.185 | 0.1988 | No |
| 22 | P2rx6 | 4460 | 0.160 | 0.1824 | No |
| 23 | Tent5c | 4573 | 0.152 | 0.1825 | No |
| 24 | Ryr1 | 4591 | 0.151 | 0.1903 | No |
| 25 | Mx2 | 4621 | 0.148 | 0.1969 | No |
| 26 | Cpeb3 | 4646 | 0.146 | 0.2038 | No |
| 27 | Kmt2d | 5269 | 0.099 | 0.1592 | No |
| 28 | Zbtb16 | 5293 | 0.097 | 0.1632 | No |
| 29 | Chst2 | 5444 | 0.087 | 0.1563 | No |
| 30 | Tg | 5629 | 0.073 | 0.1458 | No |
| 31 | Ccdc106 | 5767 | 0.064 | 0.1385 | No |
| 32 | Cyp39a1 | 5946 | 0.051 | 0.1271 | No |
| 33 | Msh5 | 6134 | 0.037 | 0.1142 | No |
| 34 | Thnsl2 | 6238 | 0.031 | 0.1077 | No |
| 35 | Ryr2 | 6319 | 0.026 | 0.1027 | No |
| 36 | Dtnb | 6374 | 0.022 | 0.0997 | No |
| 37 | Idua | 6599 | 0.006 | 0.0818 | No |
| 38 | Prkn | 6603 | 0.006 | 0.0820 | No |
| 39 | Entpd7 | 6901 | -0.010 | 0.0584 | No |
| 40 | Brdt | 6956 | -0.013 | 0.0548 | No |
| 41 | Sidt1 | 7048 | -0.019 | 0.0485 | No |
| 42 | Tcf7l1 | 7230 | -0.032 | 0.0358 | No |
| 43 | Coq8a | 7288 | -0.036 | 0.0333 | No |
| 44 | Asb7 | 7569 | -0.057 | 0.0140 | No |
| 45 | Slc29a3 | 7587 | -0.058 | 0.0162 | No |
| 46 | Sphk2 | 7776 | -0.072 | 0.0053 | No |
| 47 | Fgfr3 | 8492 | -0.125 | -0.0453 | No |
| 48 | Nr6a1 | 8564 | -0.131 | -0.0431 | No |
| 49 | Zfp112 | 8960 | -0.159 | -0.0656 | No |
| 50 | Gpr19 | 9118 | -0.172 | -0.0680 | No |
| 51 | Bard1 | 9212 | -0.180 | -0.0646 | No |
| 52 | Mthfr | 9480 | -0.201 | -0.0742 | No |
| 53 | Ptprj | 9582 | -0.209 | -0.0697 | No |
| 54 | Prodh | 9611 | -0.211 | -0.0592 | No |
| 55 | Mfsd6 | 9729 | -0.220 | -0.0553 | No |
| 56 | Mast3 | 9778 | -0.224 | -0.0457 | No |
| 57 | Selenop | 9985 | -0.246 | -0.0475 | No |
| 58 | Ntf3 | 10135 | -0.258 | -0.0440 | No |
| 59 | Vps50 | 10422 | -0.285 | -0.0499 | No |
| 60 | Plag1 | 10602 | -0.304 | -0.0461 | No |
| 61 | Cdkal1 | 10634 | -0.307 | -0.0300 | No |
| 62 | Edar | 10658 | -0.309 | -0.0131 | No |
| 63 | Itgb1bp2 | 10917 | -0.339 | -0.0136 | No |
| 64 | Thrb | 11447 | -0.414 | -0.0315 | No |
| 65 | Klk8 | 11503 | -0.424 | -0.0103 | No |
| 66 | Rsad2 | 12081 | -0.580 | -0.0221 | No |
| 67 | Tnni3 | 12271 | -0.741 | 0.0075 | No |