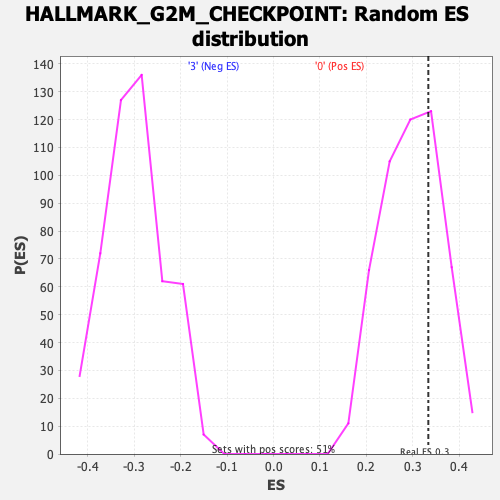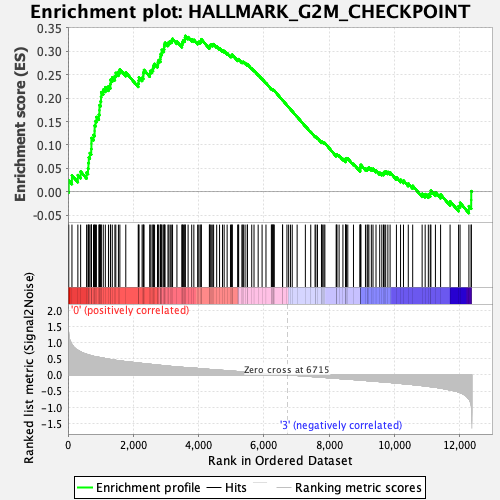
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_G2M_CHECKPOINT |
| Enrichment Score (ES) | 0.33316395 |
| Normalized Enrichment Score (NES) | 1.119613 |
| Nominal p-value | 0.2919132 |
| FDR q-value | 0.5607022 |
| FWER p-Value | 0.967 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Smc1a | 23 | 1.295 | 0.0242 | Yes |
| 2 | Pds5b | 121 | 0.941 | 0.0351 | Yes |
| 3 | Tra2b | 302 | 0.760 | 0.0357 | Yes |
| 4 | Hnrnpd | 387 | 0.713 | 0.0431 | Yes |
| 5 | Cdkn1b | 569 | 0.638 | 0.0411 | Yes |
| 6 | Kif23 | 613 | 0.624 | 0.0501 | Yes |
| 7 | Upf1 | 626 | 0.619 | 0.0616 | Yes |
| 8 | Cdc20 | 637 | 0.616 | 0.0732 | Yes |
| 9 | Egf | 669 | 0.606 | 0.0828 | Yes |
| 10 | Pttg1 | 711 | 0.595 | 0.0914 | Yes |
| 11 | Nup98 | 717 | 0.593 | 0.1030 | Yes |
| 12 | Racgap1 | 718 | 0.592 | 0.1149 | Yes |
| 13 | Tgfb1 | 781 | 0.575 | 0.1214 | Yes |
| 14 | Stil | 815 | 0.567 | 0.1301 | Yes |
| 15 | Hnrnpu | 816 | 0.567 | 0.1415 | Yes |
| 16 | Tacc3 | 842 | 0.563 | 0.1508 | Yes |
| 17 | Ube2s | 870 | 0.559 | 0.1598 | Yes |
| 18 | Slc12a2 | 945 | 0.543 | 0.1647 | Yes |
| 19 | Hmmr | 962 | 0.540 | 0.1742 | Yes |
| 20 | Rpa2 | 964 | 0.540 | 0.1850 | Yes |
| 21 | E2f3 | 997 | 0.534 | 0.1931 | Yes |
| 22 | Fbxo5 | 1011 | 0.532 | 0.2028 | Yes |
| 23 | Ttk | 1018 | 0.531 | 0.2129 | Yes |
| 24 | Orc5 | 1080 | 0.517 | 0.2183 | Yes |
| 25 | Hus1 | 1143 | 0.506 | 0.2234 | Yes |
| 26 | Ctcf | 1239 | 0.486 | 0.2254 | Yes |
| 27 | Foxn3 | 1298 | 0.477 | 0.2302 | Yes |
| 28 | Cdk1 | 1303 | 0.476 | 0.2395 | Yes |
| 29 | Mtf2 | 1356 | 0.467 | 0.2446 | Yes |
| 30 | Bub1 | 1436 | 0.454 | 0.2473 | Yes |
| 31 | Cdc45 | 1464 | 0.451 | 0.2541 | Yes |
| 32 | Top2a | 1544 | 0.438 | 0.2565 | Yes |
| 33 | Sfpq | 1588 | 0.433 | 0.2616 | Yes |
| 34 | Kif4 | 1769 | 0.412 | 0.2552 | Yes |
| 35 | Nasp | 2150 | 0.369 | 0.2314 | Yes |
| 36 | Rad54l | 2164 | 0.367 | 0.2377 | Yes |
| 37 | Odc1 | 2172 | 0.366 | 0.2445 | Yes |
| 38 | Mki67 | 2269 | 0.355 | 0.2438 | Yes |
| 39 | E2f2 | 2305 | 0.350 | 0.2480 | Yes |
| 40 | Map3k20 | 2306 | 0.350 | 0.2550 | Yes |
| 41 | Smad3 | 2328 | 0.349 | 0.2603 | Yes |
| 42 | Syncrip | 2504 | 0.329 | 0.2526 | Yes |
| 43 | Smc4 | 2516 | 0.328 | 0.2583 | Yes |
| 44 | Srsf2 | 2572 | 0.323 | 0.2602 | Yes |
| 45 | Cenpf | 2603 | 0.320 | 0.2642 | Yes |
| 46 | Ccnd1 | 2615 | 0.318 | 0.2697 | Yes |
| 47 | Dkc1 | 2650 | 0.315 | 0.2733 | Yes |
| 48 | Cul3 | 2740 | 0.305 | 0.2721 | Yes |
| 49 | Myc | 2754 | 0.303 | 0.2771 | Yes |
| 50 | Meis1 | 2776 | 0.300 | 0.2814 | Yes |
| 51 | Nup50 | 2827 | 0.296 | 0.2833 | Yes |
| 52 | Mcm6 | 2830 | 0.296 | 0.2891 | Yes |
| 53 | Smc2 | 2840 | 0.294 | 0.2942 | Yes |
| 54 | Mapk14 | 2871 | 0.292 | 0.2976 | Yes |
| 55 | Tle3 | 2873 | 0.292 | 0.3034 | Yes |
| 56 | H2az2 | 2934 | 0.287 | 0.3043 | Yes |
| 57 | Ddx39a | 2944 | 0.286 | 0.3093 | Yes |
| 58 | Katna1 | 2947 | 0.286 | 0.3149 | Yes |
| 59 | Dbf4 | 2971 | 0.282 | 0.3187 | Yes |
| 60 | Cdc27 | 3061 | 0.273 | 0.3168 | Yes |
| 61 | Cks2 | 3089 | 0.270 | 0.3200 | Yes |
| 62 | Cdc7 | 3132 | 0.265 | 0.3219 | Yes |
| 63 | Birc5 | 3177 | 0.261 | 0.3236 | Yes |
| 64 | Dr1 | 3201 | 0.259 | 0.3269 | Yes |
| 65 | Smarcc1 | 3335 | 0.248 | 0.3210 | Yes |
| 66 | Prc1 | 3488 | 0.235 | 0.3132 | Yes |
| 67 | Hmgb3 | 3495 | 0.234 | 0.3174 | Yes |
| 68 | Ccnb2 | 3510 | 0.233 | 0.3210 | Yes |
| 69 | Bcl3 | 3533 | 0.232 | 0.3238 | Yes |
| 70 | Ndc80 | 3576 | 0.227 | 0.3250 | Yes |
| 71 | Srsf1 | 3582 | 0.226 | 0.3291 | Yes |
| 72 | Tnpo2 | 3589 | 0.226 | 0.3332 | Yes |
| 73 | Bub3 | 3677 | 0.222 | 0.3305 | No |
| 74 | Cdc25a | 3787 | 0.212 | 0.3258 | No |
| 75 | Ewsr1 | 3853 | 0.209 | 0.3247 | No |
| 76 | Atrx | 3969 | 0.200 | 0.3193 | No |
| 77 | Nsd2 | 4004 | 0.197 | 0.3205 | No |
| 78 | Cenpe | 4046 | 0.194 | 0.3210 | No |
| 79 | Chek1 | 4075 | 0.192 | 0.3225 | No |
| 80 | Pola2 | 4081 | 0.191 | 0.3260 | No |
| 81 | Rad23b | 4328 | 0.169 | 0.3092 | No |
| 82 | Prim2 | 4336 | 0.169 | 0.3120 | No |
| 83 | Lig3 | 4355 | 0.167 | 0.3139 | No |
| 84 | Kif11 | 4386 | 0.165 | 0.3148 | No |
| 85 | Knl1 | 4432 | 0.162 | 0.3143 | No |
| 86 | Top1 | 4458 | 0.160 | 0.3155 | No |
| 87 | Hira | 4552 | 0.154 | 0.3110 | No |
| 88 | Kif15 | 4640 | 0.147 | 0.3068 | No |
| 89 | Rad21 | 4730 | 0.140 | 0.3023 | No |
| 90 | Incenp | 4781 | 0.137 | 0.3009 | No |
| 91 | Hif1a | 4872 | 0.128 | 0.2961 | No |
| 92 | Ilf3 | 4974 | 0.120 | 0.2902 | No |
| 93 | Nusap1 | 5003 | 0.118 | 0.2903 | No |
| 94 | H2az1 | 5025 | 0.117 | 0.2910 | No |
| 95 | Ythdc1 | 5026 | 0.117 | 0.2933 | No |
| 96 | Lbr | 5195 | 0.104 | 0.2816 | No |
| 97 | Slc38a1 | 5213 | 0.103 | 0.2823 | No |
| 98 | Amd1 | 5225 | 0.102 | 0.2834 | No |
| 99 | Mcm2 | 5324 | 0.096 | 0.2773 | No |
| 100 | Numa1 | 5351 | 0.094 | 0.2771 | No |
| 101 | Kpnb1 | 5368 | 0.092 | 0.2776 | No |
| 102 | Cdkn2c | 5421 | 0.088 | 0.2751 | No |
| 103 | Abl1 | 5490 | 0.083 | 0.2712 | No |
| 104 | Atf5 | 5492 | 0.083 | 0.2728 | No |
| 105 | Mad2l1 | 5625 | 0.073 | 0.2634 | No |
| 106 | Ncl | 5695 | 0.069 | 0.2591 | No |
| 107 | Mcm5 | 5822 | 0.060 | 0.2500 | No |
| 108 | Rasal2 | 5942 | 0.052 | 0.2413 | No |
| 109 | Ube2c | 6061 | 0.041 | 0.2324 | No |
| 110 | Cul1 | 6226 | 0.031 | 0.2196 | No |
| 111 | Mnat1 | 6242 | 0.031 | 0.2190 | No |
| 112 | Slc7a5 | 6254 | 0.030 | 0.2187 | No |
| 113 | Suv39h1 | 6276 | 0.029 | 0.2175 | No |
| 114 | Plk4 | 6278 | 0.029 | 0.2180 | No |
| 115 | Stag1 | 6300 | 0.027 | 0.2169 | No |
| 116 | Stmn1 | 6313 | 0.026 | 0.2164 | No |
| 117 | Nolc1 | 6567 | 0.008 | 0.1958 | No |
| 118 | Pura | 6571 | 0.008 | 0.1957 | No |
| 119 | Mcm3 | 6703 | 0.001 | 0.1850 | No |
| 120 | Hspa8 | 6755 | -0.000 | 0.1808 | No |
| 121 | Fancc | 6811 | -0.004 | 0.1764 | No |
| 122 | Odf2 | 6865 | -0.007 | 0.1722 | No |
| 123 | Notch2 | 7015 | -0.017 | 0.1603 | No |
| 124 | Ccna2 | 7267 | -0.035 | 0.1404 | No |
| 125 | Kif20b | 7432 | -0.047 | 0.1279 | No |
| 126 | Prpf4b | 7564 | -0.057 | 0.1183 | No |
| 127 | Gspt1 | 7589 | -0.058 | 0.1175 | No |
| 128 | Orc6 | 7638 | -0.061 | 0.1147 | No |
| 129 | Dtymk | 7760 | -0.071 | 0.1062 | No |
| 130 | Rps6ka5 | 7765 | -0.071 | 0.1073 | No |
| 131 | Kif5b | 7799 | -0.074 | 0.1061 | No |
| 132 | Pole | 7835 | -0.076 | 0.1048 | No |
| 133 | Dmd | 7864 | -0.078 | 0.1041 | No |
| 134 | Brca2 | 8210 | -0.104 | 0.0778 | No |
| 135 | Traip | 8223 | -0.105 | 0.0790 | No |
| 136 | Xpo1 | 8234 | -0.105 | 0.0802 | No |
| 137 | Prmt5 | 8300 | -0.111 | 0.0771 | No |
| 138 | Tpx2 | 8417 | -0.119 | 0.0700 | No |
| 139 | Polq | 8497 | -0.125 | 0.0661 | No |
| 140 | Jpt1 | 8500 | -0.126 | 0.0684 | No |
| 141 | Cbx1 | 8501 | -0.126 | 0.0710 | No |
| 142 | Pafah1b1 | 8524 | -0.128 | 0.0717 | No |
| 143 | Srsf10 | 8566 | -0.131 | 0.0710 | No |
| 144 | Lmnb1 | 8740 | -0.142 | 0.0597 | No |
| 145 | Sap30 | 8942 | -0.158 | 0.0463 | No |
| 146 | Wrn | 8946 | -0.158 | 0.0493 | No |
| 147 | Ss18 | 8948 | -0.158 | 0.0524 | No |
| 148 | G3bp1 | 8962 | -0.159 | 0.0545 | No |
| 149 | Exo1 | 8964 | -0.160 | 0.0576 | No |
| 150 | Mybl2 | 9107 | -0.171 | 0.0494 | No |
| 151 | Tmpo | 9158 | -0.176 | 0.0489 | No |
| 152 | Meis2 | 9182 | -0.178 | 0.0505 | No |
| 153 | Bard1 | 9212 | -0.180 | 0.0518 | No |
| 154 | Marcks | 9287 | -0.187 | 0.0495 | No |
| 155 | Aurka | 9338 | -0.189 | 0.0492 | No |
| 156 | Chaf1a | 9430 | -0.197 | 0.0457 | No |
| 157 | Slc7a1 | 9543 | -0.206 | 0.0406 | No |
| 158 | Tfdp1 | 9608 | -0.211 | 0.0396 | No |
| 159 | Uck2 | 9659 | -0.215 | 0.0398 | No |
| 160 | E2f4 | 9680 | -0.216 | 0.0425 | No |
| 161 | Cul4a | 9726 | -0.220 | 0.0433 | No |
| 162 | Ccnt1 | 9793 | -0.226 | 0.0424 | No |
| 163 | Cul5 | 9861 | -0.232 | 0.0416 | No |
| 164 | Arid4a | 10055 | -0.252 | 0.0308 | No |
| 165 | Ezh2 | 10181 | -0.262 | 0.0258 | No |
| 166 | Snrpd1 | 10271 | -0.271 | 0.0240 | No |
| 167 | Rbl1 | 10418 | -0.285 | 0.0177 | No |
| 168 | Cdc25b | 10553 | -0.298 | 0.0127 | No |
| 169 | Kmt5a | 10842 | -0.329 | -0.0043 | No |
| 170 | Casp8ap2 | 10935 | -0.341 | -0.0050 | No |
| 171 | Ccnf | 11032 | -0.354 | -0.0057 | No |
| 172 | Gins2 | 11090 | -0.360 | -0.0032 | No |
| 173 | Sqle | 11110 | -0.362 | 0.0026 | No |
| 174 | Tent4a | 11249 | -0.382 | -0.0011 | No |
| 175 | Cdc6 | 11407 | -0.407 | -0.0058 | No |
| 176 | E2f1 | 11698 | -0.463 | -0.0203 | No |
| 177 | Cenpa | 11958 | -0.530 | -0.0308 | No |
| 178 | H2ax | 12003 | -0.544 | -0.0235 | No |
| 179 | Cdk4 | 12274 | -0.745 | -0.0307 | No |
| 180 | Pml | 12344 | -0.937 | -0.0175 | No |
| 181 | Chmp1a | 12346 | -0.942 | 0.0014 | No |