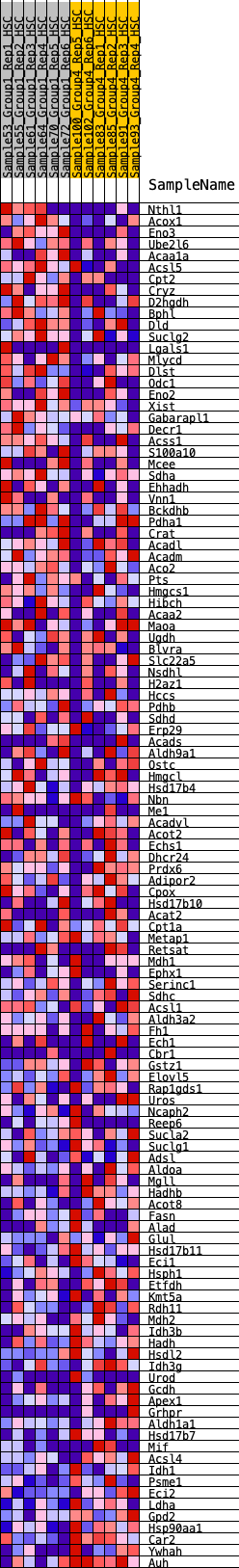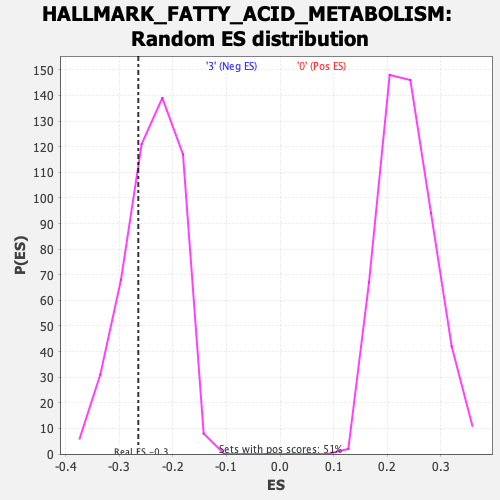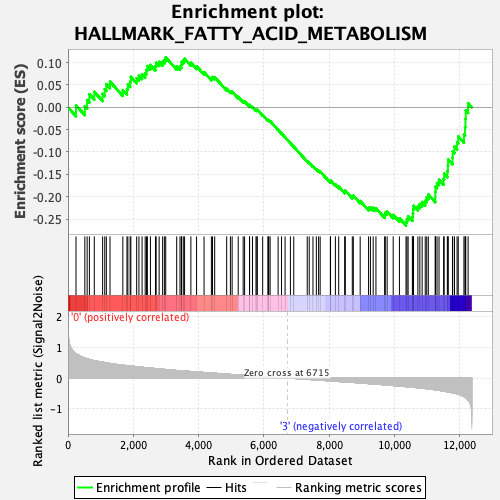
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.2647543 |
| Normalized Enrichment Score (NES) | -1.1087128 |
| Nominal p-value | 0.27142859 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nthl1 | 245 | 0.795 | 0.0044 | No |
| 2 | Acox1 | 515 | 0.654 | 0.0026 | No |
| 3 | Eno3 | 586 | 0.630 | 0.0162 | No |
| 4 | Ube2l6 | 655 | 0.610 | 0.0294 | No |
| 5 | Acaa1a | 805 | 0.570 | 0.0348 | No |
| 6 | Acsl5 | 1060 | 0.521 | 0.0301 | No |
| 7 | Cpt2 | 1125 | 0.510 | 0.0405 | No |
| 8 | Cryz | 1174 | 0.499 | 0.0519 | No |
| 9 | D2hgdh | 1286 | 0.480 | 0.0576 | No |
| 10 | Bphl | 1679 | 0.422 | 0.0386 | No |
| 11 | Dld | 1805 | 0.407 | 0.0409 | No |
| 12 | Suclg2 | 1833 | 0.404 | 0.0511 | No |
| 13 | Lgals1 | 1903 | 0.397 | 0.0577 | No |
| 14 | Mlycd | 1924 | 0.396 | 0.0682 | No |
| 15 | Dlst | 2102 | 0.375 | 0.0653 | No |
| 16 | Odc1 | 2172 | 0.366 | 0.0709 | No |
| 17 | Eno2 | 2268 | 0.355 | 0.0740 | No |
| 18 | Xist | 2363 | 0.343 | 0.0769 | No |
| 19 | Gabarapl1 | 2404 | 0.339 | 0.0841 | No |
| 20 | Decr1 | 2430 | 0.338 | 0.0924 | No |
| 21 | Acss1 | 2524 | 0.327 | 0.0949 | No |
| 22 | S100a10 | 2678 | 0.312 | 0.0920 | No |
| 23 | Mcee | 2698 | 0.310 | 0.1000 | No |
| 24 | Sdha | 2790 | 0.299 | 0.1017 | No |
| 25 | Ehhadh | 2897 | 0.290 | 0.1020 | No |
| 26 | Vnn1 | 2951 | 0.285 | 0.1064 | No |
| 27 | Bckdhb | 2992 | 0.280 | 0.1117 | No |
| 28 | Pdha1 | 3328 | 0.249 | 0.0920 | No |
| 29 | Crat | 3429 | 0.241 | 0.0913 | No |
| 30 | Acadl | 3470 | 0.236 | 0.0953 | No |
| 31 | Acadm | 3478 | 0.236 | 0.1020 | No |
| 32 | Aco2 | 3530 | 0.232 | 0.1049 | No |
| 33 | Pts | 3569 | 0.228 | 0.1088 | No |
| 34 | Hmgcs1 | 3762 | 0.215 | 0.0998 | No |
| 35 | Hibch | 3936 | 0.204 | 0.0919 | No |
| 36 | Acaa2 | 4165 | 0.183 | 0.0789 | No |
| 37 | Maoa | 4390 | 0.164 | 0.0657 | No |
| 38 | Ugdh | 4425 | 0.162 | 0.0679 | No |
| 39 | Blvra | 4491 | 0.158 | 0.0674 | No |
| 40 | Slc22a5 | 4858 | 0.129 | 0.0415 | No |
| 41 | Nsdhl | 4972 | 0.121 | 0.0360 | No |
| 42 | H2az1 | 5025 | 0.117 | 0.0353 | No |
| 43 | Hccs | 5211 | 0.103 | 0.0234 | No |
| 44 | Pdhb | 5363 | 0.093 | 0.0139 | No |
| 45 | Sdhd | 5407 | 0.089 | 0.0131 | No |
| 46 | Erp29 | 5558 | 0.078 | 0.0033 | No |
| 47 | Acads | 5559 | 0.078 | 0.0057 | No |
| 48 | Aldh9a1 | 5648 | 0.072 | 0.0007 | No |
| 49 | Ostc | 5755 | 0.065 | -0.0060 | No |
| 50 | Hmgcl | 5785 | 0.063 | -0.0064 | No |
| 51 | Hsd17b4 | 5793 | 0.062 | -0.0051 | No |
| 52 | Nbn | 5960 | 0.050 | -0.0171 | No |
| 53 | Me1 | 6119 | 0.038 | -0.0288 | No |
| 54 | Acadvl | 6142 | 0.037 | -0.0295 | No |
| 55 | Acot2 | 6144 | 0.037 | -0.0284 | No |
| 56 | Echs1 | 6188 | 0.034 | -0.0309 | No |
| 57 | Dhcr24 | 6433 | 0.018 | -0.0503 | No |
| 58 | Prdx6 | 6537 | 0.011 | -0.0584 | No |
| 59 | Adipor2 | 6646 | 0.004 | -0.0671 | No |
| 60 | Cpox | 6809 | -0.004 | -0.0802 | No |
| 61 | Hsd17b10 | 6912 | -0.010 | -0.0882 | No |
| 62 | Acat2 | 7324 | -0.039 | -0.1206 | No |
| 63 | Cpt1a | 7385 | -0.043 | -0.1241 | No |
| 64 | Metap1 | 7500 | -0.052 | -0.1318 | No |
| 65 | Retsat | 7603 | -0.059 | -0.1384 | No |
| 66 | Mdh1 | 7673 | -0.065 | -0.1420 | No |
| 67 | Ephx1 | 7720 | -0.068 | -0.1437 | No |
| 68 | Serinc1 | 8032 | -0.091 | -0.1663 | No |
| 69 | Sdhc | 8038 | -0.092 | -0.1639 | No |
| 70 | Acsl1 | 8186 | -0.102 | -0.1727 | No |
| 71 | Aldh3a2 | 8288 | -0.110 | -0.1776 | No |
| 72 | Fh1 | 8470 | -0.123 | -0.1886 | No |
| 73 | Ech1 | 8490 | -0.125 | -0.1863 | No |
| 74 | Cbr1 | 8704 | -0.140 | -0.1994 | No |
| 75 | Gstz1 | 8735 | -0.142 | -0.1975 | No |
| 76 | Elovl5 | 8945 | -0.158 | -0.2097 | No |
| 77 | Rap1gds1 | 9202 | -0.179 | -0.2251 | No |
| 78 | Uros | 9259 | -0.185 | -0.2240 | No |
| 79 | Ncaph2 | 9343 | -0.190 | -0.2249 | No |
| 80 | Reep6 | 9428 | -0.197 | -0.2257 | No |
| 81 | Sucla2 | 9694 | -0.217 | -0.2407 | No |
| 82 | Suclg1 | 9714 | -0.219 | -0.2355 | No |
| 83 | Adsl | 9771 | -0.224 | -0.2332 | No |
| 84 | Aldoa | 9956 | -0.242 | -0.2408 | No |
| 85 | Mgll | 10147 | -0.260 | -0.2483 | No |
| 86 | Hadhb | 10349 | -0.279 | -0.2562 | Yes |
| 87 | Acot8 | 10378 | -0.281 | -0.2498 | Yes |
| 88 | Fasn | 10413 | -0.285 | -0.2438 | Yes |
| 89 | Alad | 10543 | -0.297 | -0.2452 | Yes |
| 90 | Glul | 10559 | -0.299 | -0.2373 | Yes |
| 91 | Hsd17b11 | 10567 | -0.299 | -0.2287 | Yes |
| 92 | Eci1 | 10579 | -0.300 | -0.2203 | Yes |
| 93 | Hsph1 | 10712 | -0.316 | -0.2214 | Yes |
| 94 | Etfdh | 10772 | -0.323 | -0.2163 | Yes |
| 95 | Kmt5a | 10842 | -0.329 | -0.2118 | Yes |
| 96 | Rdh11 | 10937 | -0.341 | -0.2090 | Yes |
| 97 | Mdh2 | 10983 | -0.347 | -0.2020 | Yes |
| 98 | Idh3b | 11031 | -0.354 | -0.1950 | Yes |
| 99 | Hadh | 11245 | -0.381 | -0.2006 | Yes |
| 100 | Hsdl2 | 11246 | -0.381 | -0.1889 | Yes |
| 101 | Idh3g | 11252 | -0.382 | -0.1776 | Yes |
| 102 | Urod | 11298 | -0.388 | -0.1693 | Yes |
| 103 | Gcdh | 11359 | -0.398 | -0.1620 | Yes |
| 104 | Apex1 | 11500 | -0.424 | -0.1604 | Yes |
| 105 | Grhpr | 11516 | -0.426 | -0.1485 | Yes |
| 106 | Aldh1a1 | 11614 | -0.446 | -0.1427 | Yes |
| 107 | Hsd17b7 | 11632 | -0.450 | -0.1302 | Yes |
| 108 | Mif | 11638 | -0.452 | -0.1168 | Yes |
| 109 | Acsl4 | 11773 | -0.478 | -0.1130 | Yes |
| 110 | Idh1 | 11777 | -0.479 | -0.0985 | Yes |
| 111 | Psme1 | 11832 | -0.492 | -0.0878 | Yes |
| 112 | Eci2 | 11912 | -0.512 | -0.0785 | Yes |
| 113 | Ldha | 11947 | -0.525 | -0.0651 | Yes |
| 114 | Gpd2 | 12122 | -0.601 | -0.0609 | Yes |
| 115 | Hsp90aa1 | 12157 | -0.622 | -0.0445 | Yes |
| 116 | Car2 | 12167 | -0.632 | -0.0258 | Yes |
| 117 | Ywhah | 12177 | -0.638 | -0.0069 | Yes |
| 118 | Auh | 12249 | -0.717 | 0.0093 | Yes |