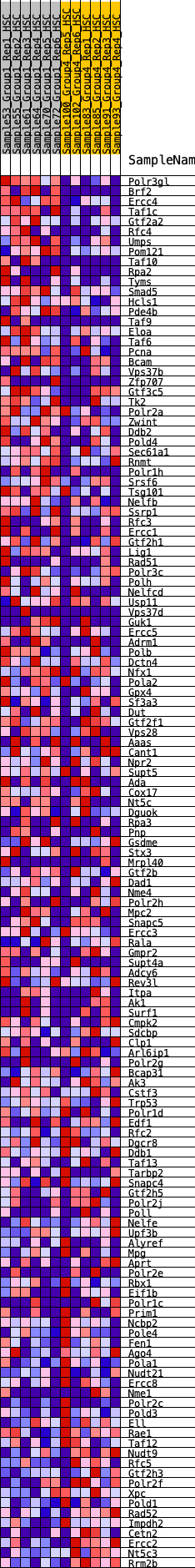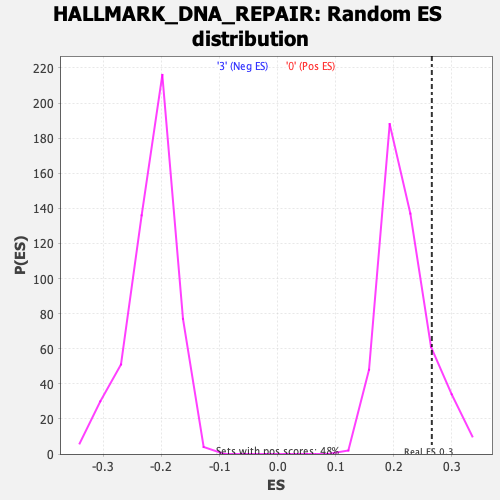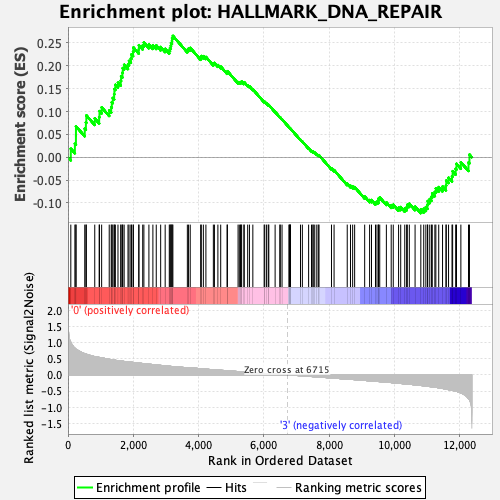
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | 0.26553467 |
| Normalized Enrichment Score (NES) | 1.2165718 |
| Nominal p-value | 0.1375 |
| FDR q-value | 0.48274562 |
| FWER p-Value | 0.897 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Polr3gl | 86 | 1.007 | 0.0187 | Yes |
| 2 | Brf2 | 211 | 0.825 | 0.0296 | Yes |
| 3 | Ercc4 | 243 | 0.796 | 0.0474 | Yes |
| 4 | Taf1c | 244 | 0.795 | 0.0677 | Yes |
| 5 | Gtf2a2 | 513 | 0.655 | 0.0625 | Yes |
| 6 | Rfc4 | 548 | 0.645 | 0.0762 | Yes |
| 7 | Umps | 562 | 0.640 | 0.0915 | Yes |
| 8 | Pom121 | 819 | 0.567 | 0.0850 | Yes |
| 9 | Taf10 | 954 | 0.541 | 0.0879 | Yes |
| 10 | Rpa2 | 964 | 0.540 | 0.1010 | Yes |
| 11 | Tyms | 1030 | 0.528 | 0.1091 | Yes |
| 12 | Smad5 | 1262 | 0.483 | 0.1025 | Yes |
| 13 | Hcls1 | 1323 | 0.473 | 0.1097 | Yes |
| 14 | Pde4b | 1341 | 0.469 | 0.1203 | Yes |
| 15 | Taf9 | 1371 | 0.465 | 0.1298 | Yes |
| 16 | Eloa | 1413 | 0.459 | 0.1382 | Yes |
| 17 | Taf6 | 1420 | 0.457 | 0.1494 | Yes |
| 18 | Pcna | 1450 | 0.452 | 0.1585 | Yes |
| 19 | Bcam | 1530 | 0.440 | 0.1633 | Yes |
| 20 | Vps37b | 1614 | 0.429 | 0.1675 | Yes |
| 21 | Zfp707 | 1626 | 0.428 | 0.1775 | Yes |
| 22 | Gtf3c5 | 1667 | 0.423 | 0.1851 | Yes |
| 23 | Tk2 | 1677 | 0.422 | 0.1951 | Yes |
| 24 | Polr2a | 1719 | 0.417 | 0.2024 | Yes |
| 25 | Zwint | 1837 | 0.403 | 0.2031 | Yes |
| 26 | Ddb2 | 1868 | 0.401 | 0.2109 | Yes |
| 27 | Pold4 | 1926 | 0.396 | 0.2164 | Yes |
| 28 | Sec61a1 | 1943 | 0.393 | 0.2251 | Yes |
| 29 | Rnmt | 1993 | 0.386 | 0.2309 | Yes |
| 30 | Polr1h | 2002 | 0.385 | 0.2401 | Yes |
| 31 | Srsf6 | 2167 | 0.367 | 0.2361 | Yes |
| 32 | Tsg101 | 2171 | 0.366 | 0.2452 | Yes |
| 33 | Nelfb | 2287 | 0.352 | 0.2448 | Yes |
| 34 | Ssrp1 | 2323 | 0.349 | 0.2508 | Yes |
| 35 | Rfc3 | 2477 | 0.332 | 0.2468 | Yes |
| 36 | Ercc1 | 2599 | 0.320 | 0.2451 | Yes |
| 37 | Gtf2h1 | 2702 | 0.310 | 0.2447 | Yes |
| 38 | Lig1 | 2837 | 0.295 | 0.2412 | Yes |
| 39 | Rad51 | 2975 | 0.281 | 0.2372 | Yes |
| 40 | Polr3c | 3101 | 0.269 | 0.2338 | Yes |
| 41 | Polh | 3123 | 0.266 | 0.2389 | Yes |
| 42 | Nelfcd | 3148 | 0.264 | 0.2437 | Yes |
| 43 | Usp11 | 3155 | 0.263 | 0.2499 | Yes |
| 44 | Vps37d | 3183 | 0.261 | 0.2544 | Yes |
| 45 | Guk1 | 3187 | 0.261 | 0.2608 | Yes |
| 46 | Ercc5 | 3211 | 0.258 | 0.2655 | Yes |
| 47 | Adrm1 | 3654 | 0.223 | 0.2351 | No |
| 48 | Polb | 3691 | 0.221 | 0.2378 | No |
| 49 | Dctn4 | 3742 | 0.217 | 0.2392 | No |
| 50 | Nfx1 | 4062 | 0.193 | 0.2181 | No |
| 51 | Pola2 | 4081 | 0.191 | 0.2215 | No |
| 52 | Gpx4 | 4144 | 0.185 | 0.2211 | No |
| 53 | Sf3a3 | 4222 | 0.179 | 0.2194 | No |
| 54 | Dut | 4450 | 0.161 | 0.2049 | No |
| 55 | Gtf2f1 | 4483 | 0.159 | 0.2064 | No |
| 56 | Vps28 | 4589 | 0.151 | 0.2016 | No |
| 57 | Aaas | 4675 | 0.143 | 0.1983 | No |
| 58 | Cant1 | 4870 | 0.128 | 0.1858 | No |
| 59 | Npr2 | 4881 | 0.127 | 0.1882 | No |
| 60 | Supt5 | 5207 | 0.104 | 0.1642 | No |
| 61 | Ada | 5245 | 0.100 | 0.1638 | No |
| 62 | Cox17 | 5282 | 0.098 | 0.1633 | No |
| 63 | Nt5c | 5297 | 0.097 | 0.1647 | No |
| 64 | Dguok | 5316 | 0.096 | 0.1657 | No |
| 65 | Rpa3 | 5386 | 0.091 | 0.1624 | No |
| 66 | Pnp | 5394 | 0.091 | 0.1641 | No |
| 67 | Gsdme | 5501 | 0.082 | 0.1575 | No |
| 68 | Stx3 | 5555 | 0.078 | 0.1552 | No |
| 69 | Mrpl40 | 5659 | 0.071 | 0.1485 | No |
| 70 | Gtf2b | 6005 | 0.045 | 0.1215 | No |
| 71 | Dad1 | 6024 | 0.044 | 0.1211 | No |
| 72 | Nme4 | 6080 | 0.041 | 0.1177 | No |
| 73 | Polr2h | 6131 | 0.037 | 0.1145 | No |
| 74 | Mpc2 | 6148 | 0.036 | 0.1142 | No |
| 75 | Snapc5 | 6341 | 0.024 | 0.0991 | No |
| 76 | Ercc3 | 6485 | 0.015 | 0.0877 | No |
| 77 | Rala | 6499 | 0.014 | 0.0870 | No |
| 78 | Gmpr2 | 6553 | 0.009 | 0.0829 | No |
| 79 | Supt4a | 6763 | -0.001 | 0.0659 | No |
| 80 | Adcy6 | 6784 | -0.002 | 0.0643 | No |
| 81 | Rev3l | 6789 | -0.002 | 0.0640 | No |
| 82 | Itpa | 6806 | -0.003 | 0.0628 | No |
| 83 | Ak1 | 6812 | -0.004 | 0.0625 | No |
| 84 | Surf1 | 7118 | -0.024 | 0.0382 | No |
| 85 | Cmpk2 | 7175 | -0.028 | 0.0343 | No |
| 86 | Sdcbp | 7367 | -0.042 | 0.0197 | No |
| 87 | Clp1 | 7455 | -0.048 | 0.0139 | No |
| 88 | Arl6ip1 | 7499 | -0.052 | 0.0117 | No |
| 89 | Polr2g | 7505 | -0.052 | 0.0126 | No |
| 90 | Bcap31 | 7554 | -0.056 | 0.0101 | No |
| 91 | Ak3 | 7617 | -0.059 | 0.0065 | No |
| 92 | Cstf3 | 7663 | -0.064 | 0.0045 | No |
| 93 | Trp53 | 7685 | -0.065 | 0.0044 | No |
| 94 | Polr1d | 8067 | -0.094 | -0.0243 | No |
| 95 | Edf1 | 8146 | -0.100 | -0.0282 | No |
| 96 | Rfc2 | 8549 | -0.130 | -0.0577 | No |
| 97 | Dgcr8 | 8649 | -0.136 | -0.0624 | No |
| 98 | Ddb1 | 8715 | -0.141 | -0.0641 | No |
| 99 | Taf13 | 8776 | -0.144 | -0.0653 | No |
| 100 | Tarbp2 | 9083 | -0.169 | -0.0860 | No |
| 101 | Snapc4 | 9234 | -0.183 | -0.0936 | No |
| 102 | Gtf2h5 | 9292 | -0.188 | -0.0935 | No |
| 103 | Polr2j | 9414 | -0.196 | -0.0984 | No |
| 104 | Poll | 9460 | -0.199 | -0.0970 | No |
| 105 | Nelfe | 9504 | -0.203 | -0.0953 | No |
| 106 | Upf3b | 9507 | -0.203 | -0.0903 | No |
| 107 | Alyref | 9544 | -0.206 | -0.0880 | No |
| 108 | Mpg | 9747 | -0.222 | -0.0988 | No |
| 109 | Aprt | 9895 | -0.235 | -0.1048 | No |
| 110 | Polr2e | 9958 | -0.243 | -0.1037 | No |
| 111 | Rbx1 | 10121 | -0.257 | -0.1104 | No |
| 112 | Eif1b | 10186 | -0.263 | -0.1089 | No |
| 113 | Polr1c | 10310 | -0.275 | -0.1119 | No |
| 114 | Prim1 | 10367 | -0.280 | -0.1094 | No |
| 115 | Ncbp2 | 10388 | -0.282 | -0.1038 | No |
| 116 | Pole4 | 10450 | -0.287 | -0.1014 | No |
| 117 | Fen1 | 10626 | -0.307 | -0.1079 | No |
| 118 | Ago4 | 10802 | -0.326 | -0.1139 | No |
| 119 | Pola1 | 10891 | -0.335 | -0.1126 | No |
| 120 | Nudt21 | 10956 | -0.343 | -0.1090 | No |
| 121 | Ercc8 | 11006 | -0.350 | -0.1041 | No |
| 122 | Nme1 | 11014 | -0.352 | -0.0957 | No |
| 123 | Polr2c | 11075 | -0.359 | -0.0914 | No |
| 124 | Pold3 | 11127 | -0.364 | -0.0863 | No |
| 125 | Ell | 11152 | -0.367 | -0.0789 | No |
| 126 | Rae1 | 11232 | -0.379 | -0.0757 | No |
| 127 | Taf12 | 11262 | -0.384 | -0.0682 | No |
| 128 | Nudt9 | 11350 | -0.396 | -0.0652 | No |
| 129 | Rfc5 | 11467 | -0.418 | -0.0640 | No |
| 130 | Gtf2h3 | 11572 | -0.438 | -0.0614 | No |
| 131 | Polr2f | 11581 | -0.440 | -0.0508 | No |
| 132 | Xpc | 11651 | -0.454 | -0.0448 | No |
| 133 | Pold1 | 11757 | -0.476 | -0.0413 | No |
| 134 | Rad52 | 11776 | -0.479 | -0.0305 | No |
| 135 | Impdh2 | 11869 | -0.499 | -0.0253 | No |
| 136 | Cetn2 | 11894 | -0.506 | -0.0143 | No |
| 137 | Ercc2 | 12028 | -0.557 | -0.0110 | No |
| 138 | Nt5c3 | 12262 | -0.731 | -0.0114 | No |
| 139 | Rrm2b | 12291 | -0.765 | 0.0059 | No |