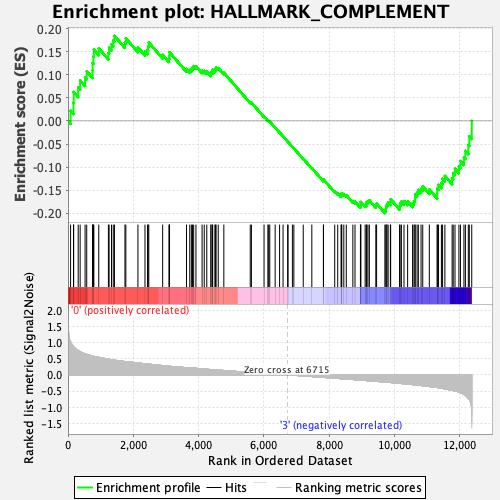
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COMPLEMENT |
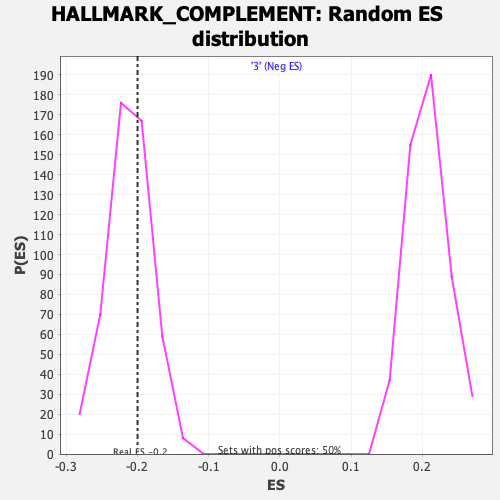
| Enrichment Score (ES) | -0.19963652 |
| Normalized Enrichment Score (NES) | -0.9473247 |
| Nominal p-value | 0.662 |
| FDR q-value | 0.9989631 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rnf4 | 79 | 1.024 | 0.0220 | No |
| 2 | Usp8 | 168 | 0.870 | 0.0391 | No |
| 3 | Grb2 | 174 | 0.865 | 0.0627 | No |
| 4 | Ehd1 | 312 | 0.756 | 0.0726 | No |
| 5 | Rhog | 372 | 0.720 | 0.0878 | No |
| 6 | Casp9 | 523 | 0.652 | 0.0937 | No |
| 7 | Jak2 | 573 | 0.637 | 0.1074 | No |
| 8 | Lrp1 | 752 | 0.584 | 0.1091 | No |
| 9 | Gnai3 | 754 | 0.583 | 0.1252 | No |
| 10 | Rabif | 779 | 0.576 | 0.1393 | No |
| 11 | F8 | 791 | 0.572 | 0.1543 | No |
| 12 | Fyn | 942 | 0.545 | 0.1572 | No |
| 13 | Pik3r5 | 1237 | 0.486 | 0.1467 | No |
| 14 | Ctsd | 1258 | 0.483 | 0.1586 | No |
| 15 | Zeb1 | 1332 | 0.471 | 0.1657 | No |
| 16 | Cr2 | 1385 | 0.463 | 0.1743 | No |
| 17 | Dgkh | 1422 | 0.457 | 0.1841 | No |
| 18 | Spock2 | 1744 | 0.415 | 0.1695 | No |
| 19 | Lta4h | 1771 | 0.411 | 0.1788 | No |
| 20 | Lck | 2141 | 0.370 | 0.1589 | No |
| 21 | Ctsl | 2357 | 0.344 | 0.1510 | No |
| 22 | Dock4 | 2436 | 0.337 | 0.1540 | No |
| 23 | Gng2 | 2452 | 0.336 | 0.1621 | No |
| 24 | Maff | 2473 | 0.333 | 0.1697 | No |
| 25 | Pik3cg | 2898 | 0.290 | 0.1431 | No |
| 26 | Pdp1 | 3091 | 0.269 | 0.1350 | No |
| 27 | Src | 3106 | 0.268 | 0.1413 | No |
| 28 | Vcpip1 | 3108 | 0.268 | 0.1487 | No |
| 29 | Gmfb | 3628 | 0.224 | 0.1125 | No |
| 30 | Adam9 | 3728 | 0.218 | 0.1105 | No |
| 31 | Mmp14 | 3777 | 0.213 | 0.1125 | No |
| 32 | Gnb4 | 3811 | 0.211 | 0.1157 | No |
| 33 | Msrb1 | 3847 | 0.209 | 0.1186 | No |
| 34 | Gp9 | 3917 | 0.204 | 0.1187 | No |
| 35 | Akap10 | 4105 | 0.189 | 0.1086 | No |
| 36 | Rbsn | 4171 | 0.183 | 0.1084 | No |
| 37 | Casp4 | 4249 | 0.176 | 0.1070 | No |
| 38 | Hspa5 | 4374 | 0.166 | 0.1015 | No |
| 39 | Plaur | 4379 | 0.165 | 0.1058 | No |
| 40 | Cpm | 4424 | 0.163 | 0.1067 | No |
| 41 | Was | 4430 | 0.162 | 0.1108 | No |
| 42 | Plek | 4497 | 0.158 | 0.1098 | No |
| 43 | Brpf3 | 4519 | 0.156 | 0.1124 | No |
| 44 | Dgkg | 4532 | 0.155 | 0.1158 | No |
| 45 | Prdm4 | 4599 | 0.150 | 0.1146 | No |
| 46 | Raf1 | 4771 | 0.137 | 0.1044 | No |
| 47 | Sirt6 | 5580 | 0.076 | 0.0405 | No |
| 48 | Cd46 | 5615 | 0.073 | 0.0398 | No |
| 49 | Ctsb | 6002 | 0.046 | 0.0095 | No |
| 50 | Me1 | 6119 | 0.038 | 0.0011 | No |
| 51 | Irf1 | 6146 | 0.036 | 0.0000 | No |
| 52 | Pclo | 6177 | 0.034 | -0.0015 | No |
| 53 | Pdgfb | 6342 | 0.024 | -0.0142 | No |
| 54 | F5 | 6478 | 0.015 | -0.0248 | No |
| 55 | Usp16 | 6584 | 0.007 | -0.0332 | No |
| 56 | C2 | 6725 | 0.000 | -0.0446 | No |
| 57 | S100a9 | 6737 | 0.000 | -0.0455 | No |
| 58 | Ppp4c | 6870 | -0.007 | -0.0561 | No |
| 59 | Atox1 | 6914 | -0.010 | -0.0593 | No |
| 60 | Gnai2 | 7202 | -0.031 | -0.0819 | No |
| 61 | Lap3 | 7465 | -0.049 | -0.1019 | No |
| 62 | Cblb | 7820 | -0.075 | -0.1288 | No |
| 63 | Ctsh | 7821 | -0.075 | -0.1267 | No |
| 64 | Gnb2 | 8166 | -0.101 | -0.1519 | No |
| 65 | Lamp2 | 8258 | -0.107 | -0.1564 | No |
| 66 | Gata3 | 8360 | -0.115 | -0.1614 | No |
| 67 | Ctsc | 8375 | -0.116 | -0.1594 | No |
| 68 | Kynu | 8381 | -0.117 | -0.1565 | No |
| 69 | Lyn | 8444 | -0.121 | -0.1582 | No |
| 70 | Dock10 | 8520 | -0.127 | -0.1608 | No |
| 71 | Plscr1 | 8721 | -0.141 | -0.1732 | No |
| 72 | Stx4a | 8787 | -0.145 | -0.1745 | No |
| 73 | Prep | 8953 | -0.159 | -0.1835 | No |
| 74 | Prss36 | 8961 | -0.159 | -0.1797 | No |
| 75 | Timp2 | 8963 | -0.160 | -0.1753 | No |
| 76 | Usp15 | 9097 | -0.170 | -0.1814 | No |
| 77 | Pim1 | 9134 | -0.173 | -0.1796 | No |
| 78 | Irf7 | 9145 | -0.174 | -0.1755 | No |
| 79 | Prkcd | 9183 | -0.178 | -0.1736 | No |
| 80 | Ctso | 9226 | -0.181 | -0.1720 | No |
| 81 | Tnfaip3 | 9424 | -0.197 | -0.1826 | No |
| 82 | Fdx1 | 9448 | -0.199 | -0.1790 | No |
| 83 | Calm3 | 9702 | -0.218 | -0.1936 | Yes |
| 84 | Psmb9 | 9735 | -0.221 | -0.1901 | Yes |
| 85 | Pik3ca | 9740 | -0.221 | -0.1842 | Yes |
| 86 | Lcp2 | 9766 | -0.223 | -0.1801 | Yes |
| 87 | Kif2a | 9801 | -0.226 | -0.1766 | Yes |
| 88 | Dock9 | 9870 | -0.233 | -0.1756 | Yes |
| 89 | Dyrk2 | 9878 | -0.234 | -0.1697 | Yes |
| 90 | Dusp6 | 10151 | -0.260 | -0.1847 | Yes |
| 91 | Prcp | 10167 | -0.262 | -0.1786 | Yes |
| 92 | Clu | 10207 | -0.265 | -0.1744 | Yes |
| 93 | Pfn1 | 10287 | -0.273 | -0.1733 | Yes |
| 94 | Xpnpep1 | 10398 | -0.283 | -0.1744 | Yes |
| 95 | Sh2b3 | 10549 | -0.298 | -0.1784 | Yes |
| 96 | Usp14 | 10592 | -0.303 | -0.1734 | Yes |
| 97 | Calm1 | 10624 | -0.307 | -0.1674 | Yes |
| 98 | Gca | 10627 | -0.307 | -0.1590 | Yes |
| 99 | Dusp5 | 10687 | -0.313 | -0.1551 | Yes |
| 100 | Casp1 | 10726 | -0.317 | -0.1494 | Yes |
| 101 | Dpp4 | 10809 | -0.327 | -0.1470 | Yes |
| 102 | Col4a2 | 10861 | -0.332 | -0.1419 | Yes |
| 103 | Csrp1 | 11062 | -0.358 | -0.1483 | Yes |
| 104 | Irf2 | 11304 | -0.389 | -0.1571 | Yes |
| 105 | S100a13 | 11305 | -0.390 | -0.1463 | Yes |
| 106 | Ppp2cb | 11344 | -0.396 | -0.1384 | Yes |
| 107 | Kcnip3 | 11432 | -0.411 | -0.1341 | Yes |
| 108 | Lipa | 11468 | -0.418 | -0.1253 | Yes |
| 109 | Ctss | 11539 | -0.430 | -0.1191 | Yes |
| 110 | Rce1 | 11760 | -0.477 | -0.1238 | Yes |
| 111 | Casp3 | 11793 | -0.483 | -0.1129 | Yes |
| 112 | Casp7 | 11848 | -0.495 | -0.1036 | Yes |
| 113 | Psen1 | 11968 | -0.533 | -0.0985 | Yes |
| 114 | Pla2g4a | 12015 | -0.549 | -0.0869 | Yes |
| 115 | Gpd2 | 12122 | -0.601 | -0.0789 | Yes |
| 116 | Car2 | 12167 | -0.632 | -0.0649 | Yes |
| 117 | Itgam | 12256 | -0.721 | -0.0520 | Yes |
| 118 | Cpq | 12286 | -0.757 | -0.0333 | Yes |
| 119 | Anxa5 | 12361 | -1.420 | 0.0002 | Yes |