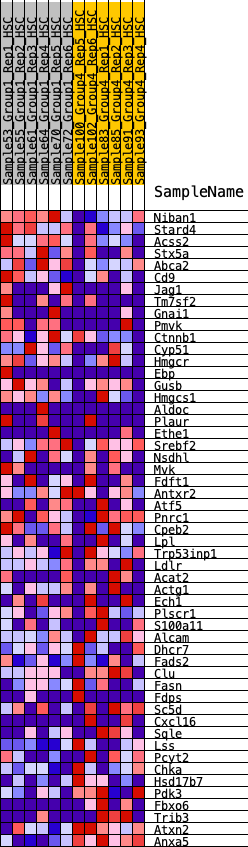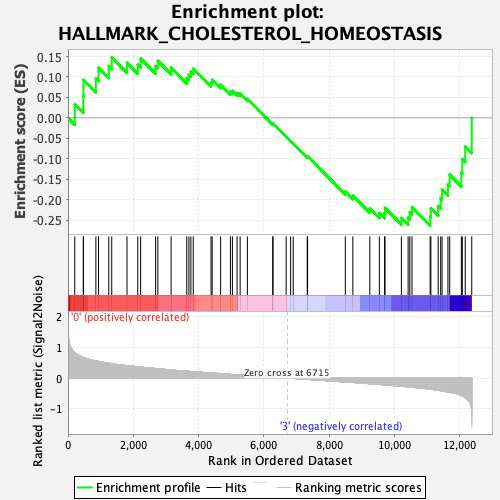
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.26273903 |
| Normalized Enrichment Score (NES) | -1.0800414 |
| Nominal p-value | 0.3548387 |
| FDR q-value | 0.8463974 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Niban1 | 207 | 0.826 | 0.0332 | No |
| 2 | Stard4 | 467 | 0.673 | 0.0529 | No |
| 3 | Acss2 | 474 | 0.671 | 0.0930 | No |
| 4 | Stx5a | 854 | 0.561 | 0.0961 | No |
| 5 | Abca2 | 935 | 0.546 | 0.1227 | No |
| 6 | Cd9 | 1249 | 0.485 | 0.1266 | No |
| 7 | Jag1 | 1339 | 0.470 | 0.1478 | No |
| 8 | Tm7sf2 | 1806 | 0.406 | 0.1345 | No |
| 9 | Gnai1 | 2136 | 0.371 | 0.1302 | No |
| 10 | Pmvk | 2224 | 0.360 | 0.1449 | No |
| 11 | Ctnnb1 | 2682 | 0.312 | 0.1267 | No |
| 12 | Cyp51 | 2751 | 0.303 | 0.1395 | No |
| 13 | Hmgcr | 3157 | 0.263 | 0.1225 | No |
| 14 | Ebp | 3636 | 0.224 | 0.0972 | No |
| 15 | Gusb | 3702 | 0.220 | 0.1052 | No |
| 16 | Hmgcs1 | 3762 | 0.215 | 0.1134 | No |
| 17 | Aldoc | 3835 | 0.209 | 0.1202 | No |
| 18 | Plaur | 4379 | 0.165 | 0.0861 | No |
| 19 | Ethe1 | 4414 | 0.163 | 0.0932 | No |
| 20 | Srebf2 | 4673 | 0.143 | 0.0810 | No |
| 21 | Nsdhl | 4972 | 0.121 | 0.0640 | No |
| 22 | Mvk | 5033 | 0.117 | 0.0662 | No |
| 23 | Fdft1 | 5176 | 0.106 | 0.0611 | No |
| 24 | Antxr2 | 5270 | 0.099 | 0.0596 | No |
| 25 | Atf5 | 5492 | 0.083 | 0.0466 | No |
| 26 | Pnrc1 | 6269 | 0.029 | -0.0147 | No |
| 27 | Cpeb2 | 6275 | 0.029 | -0.0133 | No |
| 28 | Lpl | 6679 | 0.002 | -0.0459 | No |
| 29 | Trp53inp1 | 6816 | -0.004 | -0.0567 | No |
| 30 | Ldlr | 6895 | -0.009 | -0.0625 | No |
| 31 | Acat2 | 7324 | -0.039 | -0.0949 | No |
| 32 | Actg1 | 7334 | -0.040 | -0.0932 | No |
| 33 | Ech1 | 8490 | -0.125 | -0.1795 | No |
| 34 | Plscr1 | 8721 | -0.141 | -0.1896 | No |
| 35 | S100a11 | 9240 | -0.183 | -0.2206 | No |
| 36 | Alcam | 9532 | -0.205 | -0.2319 | No |
| 37 | Dhcr7 | 9693 | -0.217 | -0.2317 | No |
| 38 | Fads2 | 9704 | -0.218 | -0.2194 | No |
| 39 | Clu | 10207 | -0.265 | -0.2441 | Yes |
| 40 | Fasn | 10413 | -0.285 | -0.2435 | Yes |
| 41 | Fdps | 10465 | -0.288 | -0.2303 | Yes |
| 42 | Sc5d | 10531 | -0.295 | -0.2177 | Yes |
| 43 | Cxcl16 | 11087 | -0.360 | -0.2409 | Yes |
| 44 | Sqle | 11110 | -0.362 | -0.2208 | Yes |
| 45 | Lss | 11334 | -0.394 | -0.2151 | Yes |
| 46 | Pcyt2 | 11410 | -0.407 | -0.1965 | Yes |
| 47 | Chka | 11453 | -0.415 | -0.1748 | Yes |
| 48 | Hsd17b7 | 11632 | -0.450 | -0.1620 | Yes |
| 49 | Pdk3 | 11686 | -0.461 | -0.1384 | Yes |
| 50 | Fbxo6 | 12043 | -0.565 | -0.1332 | Yes |
| 51 | Trib3 | 12072 | -0.576 | -0.1006 | Yes |
| 52 | Atxn2 | 12162 | -0.629 | -0.0697 | Yes |
| 53 | Anxa5 | 12361 | -1.420 | 0.0002 | Yes |