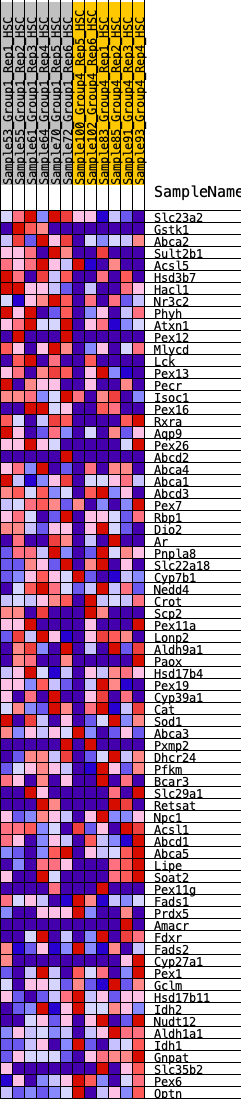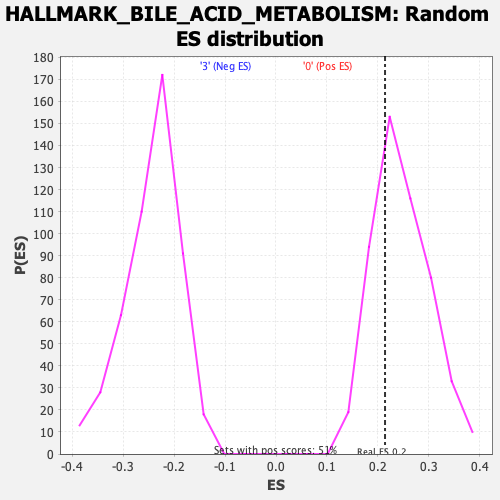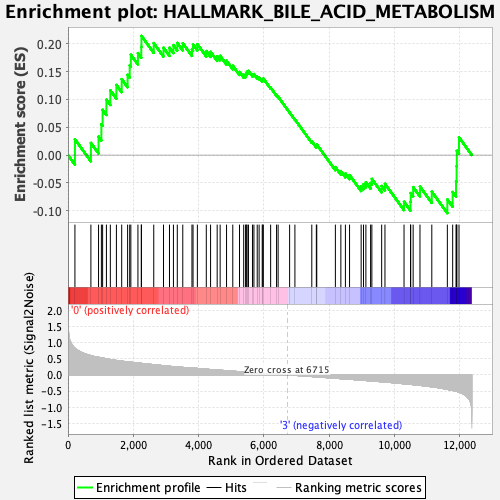
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.21424621 |
| Normalized Enrichment Score (NES) | 0.8677208 |
| Nominal p-value | 0.7029703 |
| FDR q-value | 0.9493488 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc23a2 | 212 | 0.825 | 0.0285 | Yes |
| 2 | Gstk1 | 701 | 0.598 | 0.0219 | Yes |
| 3 | Abca2 | 935 | 0.546 | 0.0331 | Yes |
| 4 | Sult2b1 | 1022 | 0.530 | 0.0555 | Yes |
| 5 | Acsl5 | 1060 | 0.521 | 0.0814 | Yes |
| 6 | Hsd3b7 | 1176 | 0.499 | 0.0996 | Yes |
| 7 | Hacl1 | 1296 | 0.477 | 0.1164 | Yes |
| 8 | Nr3c2 | 1482 | 0.447 | 0.1260 | Yes |
| 9 | Phyh | 1644 | 0.426 | 0.1365 | Yes |
| 10 | Atxn1 | 1825 | 0.405 | 0.1443 | Yes |
| 11 | Pex12 | 1889 | 0.398 | 0.1612 | Yes |
| 12 | Mlycd | 1924 | 0.396 | 0.1804 | Yes |
| 13 | Lck | 2141 | 0.370 | 0.1833 | Yes |
| 14 | Pex13 | 2244 | 0.358 | 0.1948 | Yes |
| 15 | Pecr | 2249 | 0.357 | 0.2142 | Yes |
| 16 | Isoc1 | 2625 | 0.317 | 0.2013 | No |
| 17 | Pex16 | 2924 | 0.288 | 0.1930 | No |
| 18 | Rxra | 3107 | 0.268 | 0.1930 | No |
| 19 | Aqp9 | 3228 | 0.257 | 0.1975 | No |
| 20 | Pex26 | 3346 | 0.247 | 0.2017 | No |
| 21 | Abcd2 | 3514 | 0.233 | 0.2010 | No |
| 22 | Abca4 | 3795 | 0.212 | 0.1899 | No |
| 23 | Abca1 | 3828 | 0.210 | 0.1989 | No |
| 24 | Abcd3 | 3961 | 0.201 | 0.1993 | No |
| 25 | Pex7 | 4234 | 0.177 | 0.1870 | No |
| 26 | Rbp1 | 4365 | 0.166 | 0.1856 | No |
| 27 | Dio2 | 4567 | 0.152 | 0.1777 | No |
| 28 | Ar | 4658 | 0.145 | 0.1784 | No |
| 29 | Pnpla8 | 4854 | 0.129 | 0.1697 | No |
| 30 | Slc22a18 | 5045 | 0.116 | 0.1606 | No |
| 31 | Cyp7b1 | 5251 | 0.100 | 0.1495 | No |
| 32 | Nedd4 | 5379 | 0.092 | 0.1442 | No |
| 33 | Crot | 5437 | 0.088 | 0.1445 | No |
| 34 | Scp2 | 5460 | 0.085 | 0.1474 | No |
| 35 | Pex11a | 5483 | 0.083 | 0.1502 | No |
| 36 | Lonp2 | 5525 | 0.080 | 0.1513 | No |
| 37 | Aldh9a1 | 5648 | 0.072 | 0.1453 | No |
| 38 | Paox | 5692 | 0.069 | 0.1457 | No |
| 39 | Hsd17b4 | 5793 | 0.062 | 0.1410 | No |
| 40 | Pex19 | 5851 | 0.058 | 0.1396 | No |
| 41 | Cyp39a1 | 5946 | 0.051 | 0.1348 | No |
| 42 | Cat | 5964 | 0.050 | 0.1361 | No |
| 43 | Sod1 | 5979 | 0.048 | 0.1377 | No |
| 44 | Abca3 | 6204 | 0.033 | 0.1212 | No |
| 45 | Pxmp2 | 6385 | 0.021 | 0.1077 | No |
| 46 | Dhcr24 | 6433 | 0.018 | 0.1049 | No |
| 47 | Pfkm | 6785 | -0.002 | 0.0765 | No |
| 48 | Bcar3 | 6947 | -0.013 | 0.0641 | No |
| 49 | Slc29a1 | 7464 | -0.049 | 0.0248 | No |
| 50 | Retsat | 7603 | -0.059 | 0.0168 | No |
| 51 | Npc1 | 7616 | -0.059 | 0.0191 | No |
| 52 | Acsl1 | 8186 | -0.102 | -0.0215 | No |
| 53 | Abcd1 | 8354 | -0.115 | -0.0288 | No |
| 54 | Abca5 | 8493 | -0.125 | -0.0331 | No |
| 55 | Lipe | 8620 | -0.134 | -0.0359 | No |
| 56 | Soat2 | 8975 | -0.160 | -0.0558 | No |
| 57 | Pex11g | 9047 | -0.166 | -0.0524 | No |
| 58 | Fads1 | 9120 | -0.172 | -0.0487 | No |
| 59 | Prdx5 | 9264 | -0.185 | -0.0501 | No |
| 60 | Amacr | 9303 | -0.188 | -0.0428 | No |
| 61 | Fdxr | 9603 | -0.211 | -0.0555 | No |
| 62 | Fads2 | 9704 | -0.218 | -0.0515 | No |
| 63 | Cyp27a1 | 10288 | -0.273 | -0.0839 | No |
| 64 | Pex1 | 10486 | -0.290 | -0.0838 | No |
| 65 | Gclm | 10493 | -0.291 | -0.0682 | No |
| 66 | Hsd17b11 | 10567 | -0.299 | -0.0576 | No |
| 67 | Idh2 | 10776 | -0.323 | -0.0566 | No |
| 68 | Nudt12 | 11138 | -0.365 | -0.0657 | No |
| 69 | Aldh1a1 | 11614 | -0.446 | -0.0797 | No |
| 70 | Idh1 | 11777 | -0.479 | -0.0663 | No |
| 71 | Gnpat | 11880 | -0.503 | -0.0468 | No |
| 72 | Slc35b2 | 11898 | -0.507 | -0.0201 | No |
| 73 | Pex6 | 11904 | -0.508 | 0.0077 | No |
| 74 | Optn | 11971 | -0.534 | 0.0319 | No |