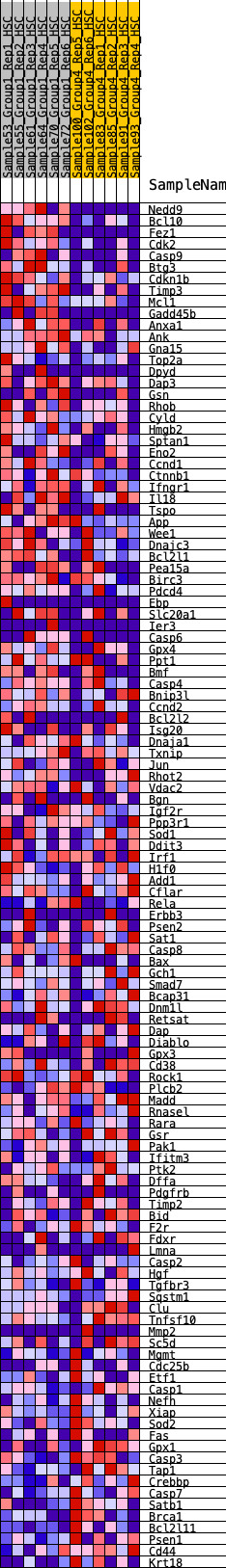
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APOPTOSIS |
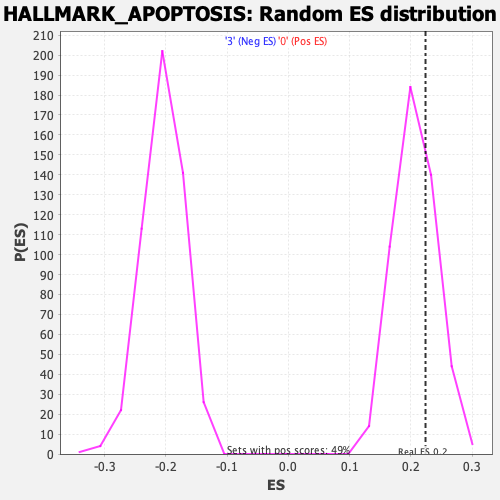
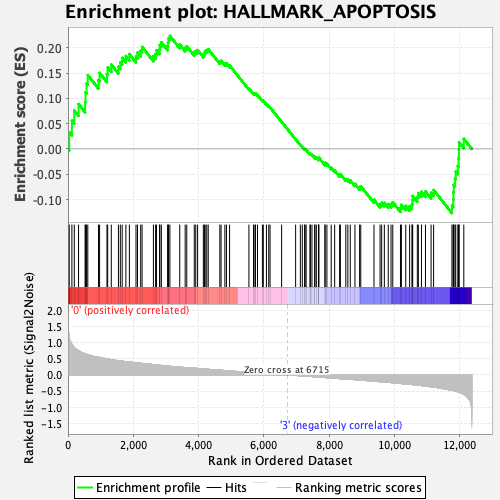
| Enrichment Score (ES) | 0.22370954 |
| Normalized Enrichment Score (NES) | 1.0822252 |
| Nominal p-value | 0.28105906 |
| FDR q-value | 0.5797482 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nedd9 | 38 | 1.162 | 0.0331 | Yes |
| 2 | Bcl10 | 119 | 0.942 | 0.0559 | Yes |
| 3 | Fez1 | 192 | 0.836 | 0.0761 | Yes |
| 4 | Cdk2 | 323 | 0.751 | 0.0889 | Yes |
| 5 | Casp9 | 523 | 0.652 | 0.0929 | Yes |
| 6 | Btg3 | 537 | 0.650 | 0.1121 | Yes |
| 7 | Cdkn1b | 569 | 0.638 | 0.1295 | Yes |
| 8 | Timp3 | 606 | 0.626 | 0.1460 | Yes |
| 9 | Mcl1 | 933 | 0.546 | 0.1364 | Yes |
| 10 | Gadd45b | 967 | 0.540 | 0.1505 | Yes |
| 11 | Anxa1 | 1193 | 0.496 | 0.1476 | Yes |
| 12 | Ank | 1215 | 0.491 | 0.1612 | Yes |
| 13 | Gna15 | 1325 | 0.472 | 0.1670 | Yes |
| 14 | Top2a | 1544 | 0.438 | 0.1628 | Yes |
| 15 | Dpyd | 1602 | 0.431 | 0.1716 | Yes |
| 16 | Dap3 | 1659 | 0.424 | 0.1802 | Yes |
| 17 | Gsn | 1772 | 0.411 | 0.1839 | Yes |
| 18 | Rhob | 1879 | 0.400 | 0.1877 | Yes |
| 19 | Cyld | 2082 | 0.376 | 0.1829 | Yes |
| 20 | Hmgb2 | 2128 | 0.372 | 0.1908 | Yes |
| 21 | Sptan1 | 2225 | 0.360 | 0.1942 | Yes |
| 22 | Eno2 | 2268 | 0.355 | 0.2018 | Yes |
| 23 | Ccnd1 | 2615 | 0.318 | 0.1835 | Yes |
| 24 | Ctnnb1 | 2682 | 0.312 | 0.1878 | Yes |
| 25 | Ifngr1 | 2710 | 0.309 | 0.1952 | Yes |
| 26 | Il18 | 2805 | 0.298 | 0.1968 | Yes |
| 27 | Tspo | 2811 | 0.297 | 0.2057 | Yes |
| 28 | App | 2858 | 0.293 | 0.2110 | Yes |
| 29 | Wee1 | 3050 | 0.273 | 0.2040 | Yes |
| 30 | Dnajc3 | 3064 | 0.272 | 0.2114 | Yes |
| 31 | Bcl2l1 | 3078 | 0.271 | 0.2188 | Yes |
| 32 | Pea15a | 3120 | 0.267 | 0.2237 | Yes |
| 33 | Birc3 | 3421 | 0.242 | 0.2067 | No |
| 34 | Pdcd4 | 3587 | 0.226 | 0.2003 | No |
| 35 | Ebp | 3636 | 0.224 | 0.2034 | No |
| 36 | Slc20a1 | 3866 | 0.208 | 0.1911 | No |
| 37 | Ier3 | 3911 | 0.204 | 0.1939 | No |
| 38 | Casp6 | 3968 | 0.201 | 0.1956 | No |
| 39 | Gpx4 | 4144 | 0.185 | 0.1871 | No |
| 40 | Ppt1 | 4176 | 0.182 | 0.1902 | No |
| 41 | Bmf | 4200 | 0.180 | 0.1939 | No |
| 42 | Casp4 | 4249 | 0.176 | 0.1955 | No |
| 43 | Bnip3l | 4293 | 0.172 | 0.1974 | No |
| 44 | Ccnd2 | 4649 | 0.146 | 0.1729 | No |
| 45 | Bcl2l2 | 4686 | 0.143 | 0.1744 | No |
| 46 | Isg20 | 4803 | 0.134 | 0.1691 | No |
| 47 | Dnaja1 | 4850 | 0.130 | 0.1694 | No |
| 48 | Txnip | 4947 | 0.123 | 0.1654 | No |
| 49 | Jun | 5539 | 0.079 | 0.1196 | No |
| 50 | Rhot2 | 5679 | 0.070 | 0.1105 | No |
| 51 | Vdac2 | 5723 | 0.067 | 0.1090 | No |
| 52 | Bgn | 5739 | 0.066 | 0.1099 | No |
| 53 | Igf2r | 5802 | 0.061 | 0.1067 | No |
| 54 | Ppp3r1 | 5952 | 0.051 | 0.0961 | No |
| 55 | Sod1 | 5979 | 0.048 | 0.0955 | No |
| 56 | Ddit3 | 6075 | 0.041 | 0.0890 | No |
| 57 | Irf1 | 6146 | 0.036 | 0.0845 | No |
| 58 | H1f0 | 6186 | 0.034 | 0.0823 | No |
| 59 | Add1 | 6540 | 0.010 | 0.0538 | No |
| 60 | Cflar | 6969 | -0.014 | 0.0193 | No |
| 61 | Rela | 7113 | -0.024 | 0.0084 | No |
| 62 | Erbb3 | 7173 | -0.028 | 0.0044 | No |
| 63 | Psen2 | 7244 | -0.034 | -0.0002 | No |
| 64 | Sat1 | 7257 | -0.034 | -0.0001 | No |
| 65 | Casp8 | 7291 | -0.037 | -0.0017 | No |
| 66 | Bax | 7409 | -0.045 | -0.0098 | No |
| 67 | Gch1 | 7420 | -0.046 | -0.0092 | No |
| 68 | Smad7 | 7469 | -0.049 | -0.0116 | No |
| 69 | Bcap31 | 7554 | -0.056 | -0.0167 | No |
| 70 | Dnm1l | 7558 | -0.056 | -0.0152 | No |
| 71 | Retsat | 7603 | -0.059 | -0.0170 | No |
| 72 | Dap | 7668 | -0.064 | -0.0202 | No |
| 73 | Diablo | 7671 | -0.064 | -0.0184 | No |
| 74 | Gpx3 | 7682 | -0.065 | -0.0172 | No |
| 75 | Cd38 | 7868 | -0.079 | -0.0298 | No |
| 76 | Rock1 | 7869 | -0.079 | -0.0274 | No |
| 77 | Plcb2 | 7927 | -0.083 | -0.0295 | No |
| 78 | Madd | 8058 | -0.094 | -0.0371 | No |
| 79 | Rnasel | 8164 | -0.101 | -0.0426 | No |
| 80 | Rara | 8316 | -0.112 | -0.0514 | No |
| 81 | Gsr | 8340 | -0.114 | -0.0498 | No |
| 82 | Pak1 | 8506 | -0.126 | -0.0593 | No |
| 83 | Ifitm3 | 8568 | -0.131 | -0.0602 | No |
| 84 | Ptk2 | 8642 | -0.135 | -0.0619 | No |
| 85 | Dffa | 8785 | -0.145 | -0.0690 | No |
| 86 | Pdgfrb | 8928 | -0.156 | -0.0758 | No |
| 87 | Timp2 | 8963 | -0.160 | -0.0736 | No |
| 88 | Bid | 9368 | -0.192 | -0.1006 | No |
| 89 | F2r | 9554 | -0.207 | -0.1092 | No |
| 90 | Fdxr | 9603 | -0.211 | -0.1066 | No |
| 91 | Lmna | 9685 | -0.216 | -0.1065 | No |
| 92 | Casp2 | 9805 | -0.227 | -0.1091 | No |
| 93 | Hgf | 9890 | -0.235 | -0.1087 | No |
| 94 | Tgfbr3 | 9943 | -0.241 | -0.1054 | No |
| 95 | Sqstm1 | 10179 | -0.262 | -0.1164 | No |
| 96 | Clu | 10207 | -0.265 | -0.1104 | No |
| 97 | Tnfsf10 | 10341 | -0.277 | -0.1126 | No |
| 98 | Mmp2 | 10459 | -0.288 | -0.1132 | No |
| 99 | Sc5d | 10531 | -0.295 | -0.1098 | No |
| 100 | Mgmt | 10545 | -0.297 | -0.1016 | No |
| 101 | Cdc25b | 10553 | -0.298 | -0.0929 | No |
| 102 | Etf1 | 10695 | -0.314 | -0.0946 | No |
| 103 | Casp1 | 10726 | -0.317 | -0.0871 | No |
| 104 | Nefh | 10821 | -0.328 | -0.0846 | No |
| 105 | Xiap | 10944 | -0.341 | -0.0839 | No |
| 106 | Sod2 | 11116 | -0.363 | -0.0866 | No |
| 107 | Fas | 11193 | -0.374 | -0.0812 | No |
| 108 | Gpx1 | 11755 | -0.476 | -0.1121 | No |
| 109 | Casp3 | 11793 | -0.483 | -0.1001 | No |
| 110 | Tap1 | 11800 | -0.485 | -0.0855 | No |
| 111 | Crebbp | 11810 | -0.488 | -0.0710 | No |
| 112 | Casp7 | 11848 | -0.495 | -0.0586 | No |
| 113 | Satb1 | 11871 | -0.500 | -0.0448 | No |
| 114 | Brca1 | 11935 | -0.521 | -0.0337 | No |
| 115 | Bcl2l11 | 11957 | -0.530 | -0.0189 | No |
| 116 | Psen1 | 11968 | -0.533 | -0.0031 | No |
| 117 | Cd44 | 11972 | -0.534 | 0.0133 | No |
| 118 | Krt18 | 12118 | -0.596 | 0.0200 | No |