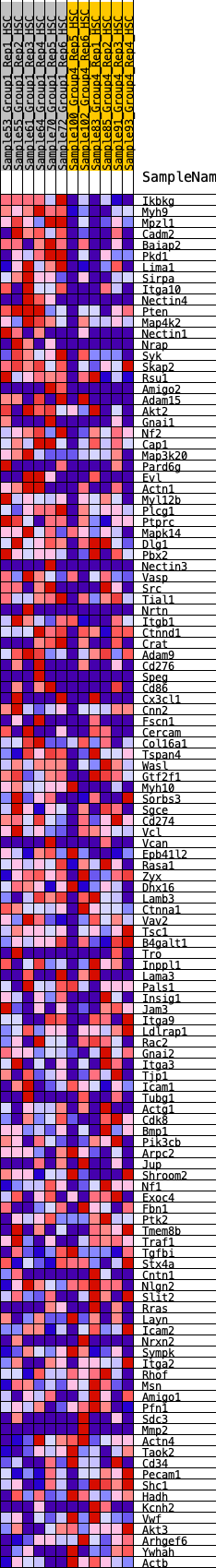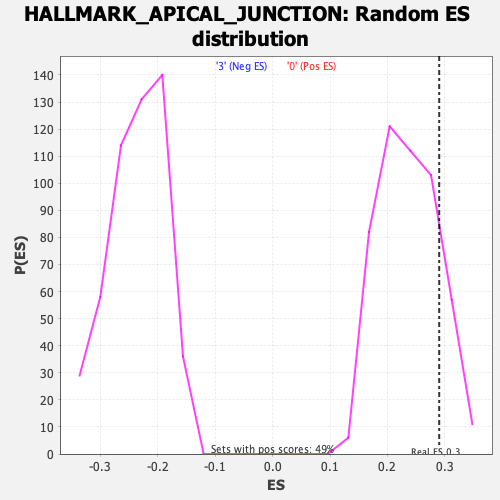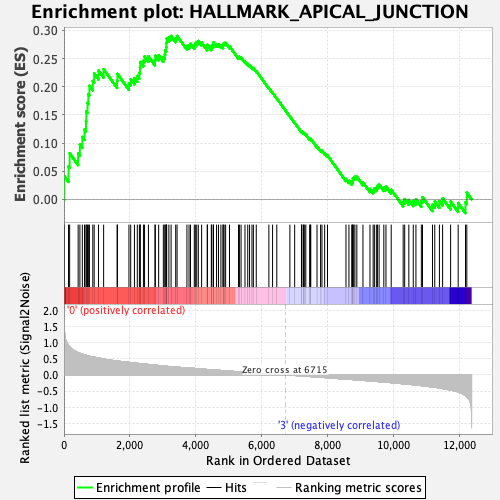
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group4.HSC_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.29028663 |
| Normalized Enrichment Score (NES) | 1.2324148 |
| Nominal p-value | 0.15447155 |
| FDR q-value | 0.5810183 |
| FWER p-Value | 0.89 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ikbkg | 10 | 1.395 | 0.0412 | Yes |
| 2 | Myh9 | 137 | 0.913 | 0.0584 | Yes |
| 3 | Mpzl1 | 171 | 0.865 | 0.0818 | Yes |
| 4 | Cadm2 | 429 | 0.688 | 0.0816 | Yes |
| 5 | Baiap2 | 482 | 0.668 | 0.0974 | Yes |
| 6 | Pkd1 | 556 | 0.642 | 0.1108 | Yes |
| 7 | Lima1 | 622 | 0.620 | 0.1242 | Yes |
| 8 | Sirpa | 667 | 0.606 | 0.1389 | Yes |
| 9 | Itga10 | 677 | 0.604 | 0.1563 | Yes |
| 10 | Nectin4 | 714 | 0.593 | 0.1713 | Yes |
| 11 | Pten | 739 | 0.587 | 0.1870 | Yes |
| 12 | Map4k2 | 771 | 0.577 | 0.2018 | Yes |
| 13 | Nectin1 | 874 | 0.558 | 0.2103 | Yes |
| 14 | Nrap | 915 | 0.550 | 0.2236 | Yes |
| 15 | Syk | 1047 | 0.524 | 0.2287 | Yes |
| 16 | Skap2 | 1202 | 0.494 | 0.2310 | Yes |
| 17 | Rsu1 | 1611 | 0.430 | 0.2107 | Yes |
| 18 | Amigo2 | 1620 | 0.428 | 0.2229 | Yes |
| 19 | Adam15 | 1970 | 0.389 | 0.2061 | Yes |
| 20 | Akt2 | 2023 | 0.383 | 0.2134 | Yes |
| 21 | Gnai1 | 2136 | 0.371 | 0.2154 | Yes |
| 22 | Nf2 | 2223 | 0.360 | 0.2193 | Yes |
| 23 | Cap1 | 2286 | 0.352 | 0.2248 | Yes |
| 24 | Map3k20 | 2306 | 0.350 | 0.2338 | Yes |
| 25 | Pard6g | 2313 | 0.350 | 0.2439 | Yes |
| 26 | Evl | 2409 | 0.339 | 0.2463 | Yes |
| 27 | Actn1 | 2443 | 0.336 | 0.2538 | Yes |
| 28 | Myl12b | 2561 | 0.324 | 0.2540 | Yes |
| 29 | Plcg1 | 2759 | 0.302 | 0.2470 | Yes |
| 30 | Ptprc | 2770 | 0.301 | 0.2552 | Yes |
| 31 | Mapk14 | 2871 | 0.292 | 0.2558 | Yes |
| 32 | Dlg1 | 3013 | 0.278 | 0.2527 | Yes |
| 33 | Pbx2 | 3058 | 0.273 | 0.2573 | Yes |
| 34 | Nectin3 | 3069 | 0.272 | 0.2647 | Yes |
| 35 | Vasp | 3100 | 0.269 | 0.2704 | Yes |
| 36 | Src | 3106 | 0.268 | 0.2780 | Yes |
| 37 | Tial1 | 3114 | 0.267 | 0.2855 | Yes |
| 38 | Nrtn | 3181 | 0.261 | 0.2880 | Yes |
| 39 | Itgb1 | 3248 | 0.255 | 0.2903 | Yes |
| 40 | Ctnnd1 | 3388 | 0.245 | 0.2863 | Yes |
| 41 | Crat | 3429 | 0.241 | 0.2903 | Yes |
| 42 | Adam9 | 3728 | 0.218 | 0.2725 | No |
| 43 | Cd276 | 3792 | 0.212 | 0.2738 | No |
| 44 | Speg | 3837 | 0.209 | 0.2765 | No |
| 45 | Cd86 | 3952 | 0.202 | 0.2732 | No |
| 46 | Cx3cl1 | 3983 | 0.199 | 0.2768 | No |
| 47 | Cnn2 | 4025 | 0.196 | 0.2793 | No |
| 48 | Fscn1 | 4073 | 0.192 | 0.2813 | No |
| 49 | Cercam | 4174 | 0.182 | 0.2786 | No |
| 50 | Col16a1 | 4346 | 0.168 | 0.2697 | No |
| 51 | Tspan4 | 4348 | 0.168 | 0.2747 | No |
| 52 | Wasl | 4468 | 0.160 | 0.2697 | No |
| 53 | Gtf2f1 | 4483 | 0.159 | 0.2734 | No |
| 54 | Myh10 | 4530 | 0.156 | 0.2743 | No |
| 55 | Sorbs3 | 4534 | 0.155 | 0.2787 | No |
| 56 | Sgce | 4627 | 0.148 | 0.2757 | No |
| 57 | Cd274 | 4685 | 0.143 | 0.2753 | No |
| 58 | Vcl | 4759 | 0.138 | 0.2735 | No |
| 59 | Vcan | 4824 | 0.133 | 0.2723 | No |
| 60 | Epb41l2 | 4826 | 0.133 | 0.2762 | No |
| 61 | Rasa1 | 4856 | 0.129 | 0.2777 | No |
| 62 | Zyx | 4902 | 0.126 | 0.2778 | No |
| 63 | Dhx16 | 5020 | 0.118 | 0.2718 | No |
| 64 | Lamb3 | 5292 | 0.098 | 0.2526 | No |
| 65 | Ctnna1 | 5320 | 0.096 | 0.2533 | No |
| 66 | Vav2 | 5370 | 0.092 | 0.2521 | No |
| 67 | Tsc1 | 5489 | 0.083 | 0.2450 | No |
| 68 | B4galt1 | 5573 | 0.077 | 0.2405 | No |
| 69 | Tro | 5628 | 0.073 | 0.2383 | No |
| 70 | Inppl1 | 5701 | 0.068 | 0.2344 | No |
| 71 | Lama3 | 5744 | 0.066 | 0.2330 | No |
| 72 | Pals1 | 5831 | 0.060 | 0.2278 | No |
| 73 | Insig1 | 6214 | 0.032 | 0.1975 | No |
| 74 | Jam3 | 6326 | 0.026 | 0.1892 | No |
| 75 | Itga9 | 6454 | 0.016 | 0.1794 | No |
| 76 | Ldlrap1 | 6851 | -0.006 | 0.1472 | No |
| 77 | Rac2 | 6996 | -0.016 | 0.1359 | No |
| 78 | Gnai2 | 7202 | -0.031 | 0.1201 | No |
| 79 | Itga3 | 7207 | -0.031 | 0.1207 | No |
| 80 | Tjp1 | 7254 | -0.034 | 0.1180 | No |
| 81 | Icam1 | 7262 | -0.035 | 0.1185 | No |
| 82 | Tubg1 | 7300 | -0.037 | 0.1165 | No |
| 83 | Actg1 | 7334 | -0.040 | 0.1151 | No |
| 84 | Cdk8 | 7450 | -0.048 | 0.1071 | No |
| 85 | Bmp1 | 7477 | -0.050 | 0.1065 | No |
| 86 | Pik3cb | 7484 | -0.051 | 0.1075 | No |
| 87 | Arpc2 | 7674 | -0.065 | 0.0940 | No |
| 88 | Jup | 7782 | -0.072 | 0.0875 | No |
| 89 | Shroom2 | 7824 | -0.076 | 0.0864 | No |
| 90 | Nf1 | 7906 | -0.081 | 0.0822 | No |
| 91 | Exoc4 | 7992 | -0.088 | 0.0779 | No |
| 92 | Fbn1 | 8552 | -0.130 | 0.0362 | No |
| 93 | Ptk2 | 8642 | -0.135 | 0.0330 | No |
| 94 | Tmem8b | 8727 | -0.141 | 0.0304 | No |
| 95 | Traf1 | 8751 | -0.143 | 0.0328 | No |
| 96 | Tgfbi | 8761 | -0.143 | 0.0364 | No |
| 97 | Stx4a | 8787 | -0.145 | 0.0387 | No |
| 98 | Cntn1 | 8823 | -0.148 | 0.0403 | No |
| 99 | Nlgn2 | 8882 | -0.152 | 0.0402 | No |
| 100 | Slit2 | 9067 | -0.168 | 0.0302 | No |
| 101 | Rras | 9280 | -0.186 | 0.0185 | No |
| 102 | Layn | 9376 | -0.193 | 0.0166 | No |
| 103 | Icam2 | 9413 | -0.196 | 0.0195 | No |
| 104 | Nrxn2 | 9473 | -0.200 | 0.0207 | No |
| 105 | Sympk | 9506 | -0.203 | 0.0242 | No |
| 106 | Itga2 | 9557 | -0.207 | 0.0264 | No |
| 107 | Rhof | 9700 | -0.217 | 0.0214 | No |
| 108 | Msn | 9765 | -0.223 | 0.0228 | No |
| 109 | Amigo1 | 9924 | -0.239 | 0.0171 | No |
| 110 | Pfn1 | 10287 | -0.273 | -0.0042 | No |
| 111 | Sdc3 | 10333 | -0.277 | 0.0005 | No |
| 112 | Mmp2 | 10459 | -0.288 | -0.0011 | No |
| 113 | Actn4 | 10593 | -0.303 | -0.0028 | No |
| 114 | Taok2 | 10674 | -0.313 | 0.0001 | No |
| 115 | Cd34 | 10838 | -0.329 | -0.0034 | No |
| 116 | Pecam1 | 10873 | -0.333 | 0.0039 | No |
| 117 | Shc1 | 11176 | -0.372 | -0.0096 | No |
| 118 | Hadh | 11245 | -0.381 | -0.0036 | No |
| 119 | Kcnh2 | 11386 | -0.402 | -0.0030 | No |
| 120 | Vwf | 11483 | -0.420 | 0.0019 | No |
| 121 | Akt3 | 11726 | -0.470 | -0.0038 | No |
| 122 | Arhgef6 | 11955 | -0.530 | -0.0064 | No |
| 123 | Ywhah | 12177 | -0.638 | -0.0052 | No |
| 124 | Actb | 12217 | -0.676 | 0.0119 | No |