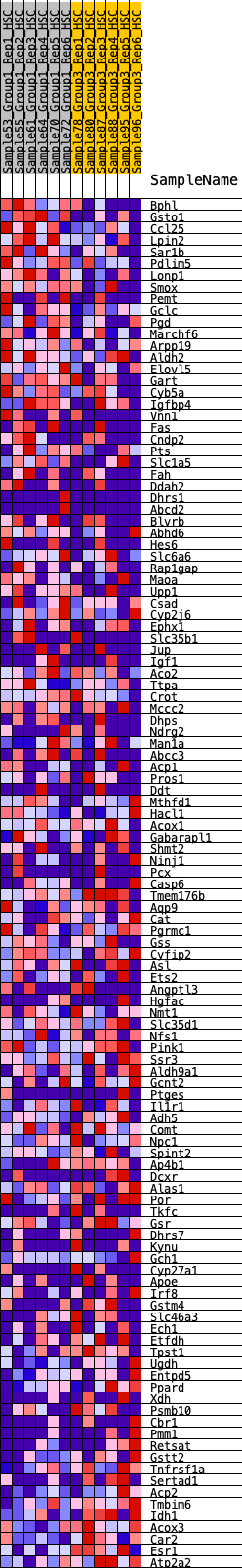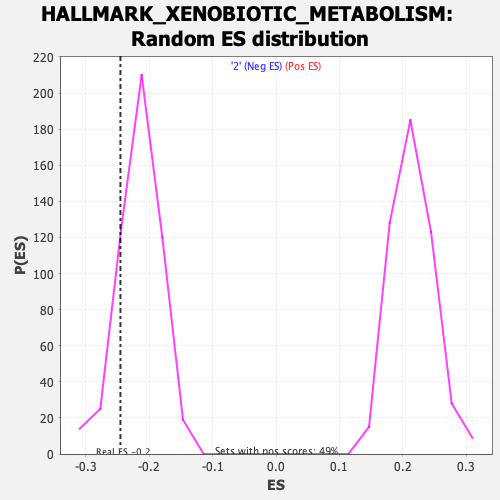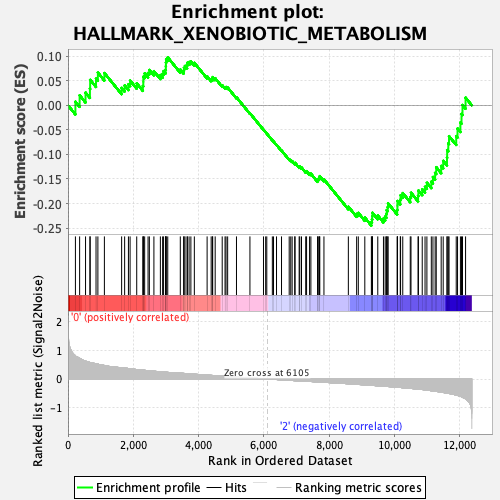
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.24505578 |
| Normalized Enrichment Score (NES) | -1.1343706 |
| Nominal p-value | 0.17773438 |
| FDR q-value | 0.80722296 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bphl | 227 | 0.794 | 0.0074 | No |
| 2 | Gsto1 | 356 | 0.715 | 0.0204 | No |
| 3 | Ccl25 | 537 | 0.618 | 0.0259 | No |
| 4 | Lpin2 | 671 | 0.573 | 0.0338 | No |
| 5 | Sar1b | 679 | 0.571 | 0.0519 | No |
| 6 | Pdlim5 | 858 | 0.526 | 0.0546 | No |
| 7 | Lonp1 | 915 | 0.510 | 0.0667 | No |
| 8 | Smox | 1114 | 0.467 | 0.0658 | No |
| 9 | Pemt | 1642 | 0.390 | 0.0355 | No |
| 10 | Gclc | 1734 | 0.378 | 0.0405 | No |
| 11 | Pgd | 1850 | 0.363 | 0.0430 | No |
| 12 | Marchf6 | 1901 | 0.356 | 0.0505 | No |
| 13 | Arpp19 | 2104 | 0.327 | 0.0447 | No |
| 14 | Aldh2 | 2292 | 0.308 | 0.0395 | No |
| 15 | Elovl5 | 2306 | 0.305 | 0.0485 | No |
| 16 | Gart | 2307 | 0.305 | 0.0584 | No |
| 17 | Cyb5a | 2347 | 0.300 | 0.0651 | No |
| 18 | Igfbp4 | 2451 | 0.287 | 0.0661 | No |
| 19 | Vnn1 | 2491 | 0.281 | 0.0721 | No |
| 20 | Fas | 2630 | 0.269 | 0.0696 | No |
| 21 | Cndp2 | 2828 | 0.251 | 0.0617 | No |
| 22 | Pts | 2897 | 0.243 | 0.0641 | No |
| 23 | Slc1a5 | 2926 | 0.241 | 0.0697 | No |
| 24 | Fah | 2988 | 0.234 | 0.0724 | No |
| 25 | Ddah2 | 2995 | 0.233 | 0.0795 | No |
| 26 | Dhrs1 | 2998 | 0.233 | 0.0870 | No |
| 27 | Abcd2 | 3005 | 0.233 | 0.0941 | No |
| 28 | Blvrb | 3054 | 0.229 | 0.0977 | No |
| 29 | Abhd6 | 3438 | 0.202 | 0.0730 | No |
| 30 | Hes6 | 3541 | 0.194 | 0.0710 | No |
| 31 | Slc6a6 | 3551 | 0.193 | 0.0766 | No |
| 32 | Rap1gap | 3582 | 0.190 | 0.0804 | No |
| 33 | Maoa | 3646 | 0.186 | 0.0813 | No |
| 34 | Upp1 | 3654 | 0.185 | 0.0868 | No |
| 35 | Csad | 3707 | 0.180 | 0.0884 | No |
| 36 | Cyp2j6 | 3762 | 0.175 | 0.0897 | No |
| 37 | Ephx1 | 3868 | 0.165 | 0.0866 | No |
| 38 | Slc35b1 | 4259 | 0.134 | 0.0591 | No |
| 39 | Jup | 4383 | 0.125 | 0.0532 | No |
| 40 | Igf1 | 4422 | 0.123 | 0.0541 | No |
| 41 | Aco2 | 4430 | 0.122 | 0.0575 | No |
| 42 | Ttpa | 4506 | 0.116 | 0.0552 | No |
| 43 | Crot | 4721 | 0.097 | 0.0409 | No |
| 44 | Mccc2 | 4808 | 0.091 | 0.0368 | No |
| 45 | Dhps | 4838 | 0.089 | 0.0374 | No |
| 46 | Ndrg2 | 4888 | 0.085 | 0.0362 | No |
| 47 | Man1a | 5158 | 0.065 | 0.0163 | No |
| 48 | Abcc3 | 5570 | 0.036 | -0.0161 | No |
| 49 | Acp1 | 5985 | 0.008 | -0.0496 | No |
| 50 | Pros1 | 6052 | 0.003 | -0.0549 | No |
| 51 | Ddt | 6086 | 0.001 | -0.0576 | No |
| 52 | Mthfd1 | 6256 | -0.008 | -0.0711 | No |
| 53 | Hacl1 | 6292 | -0.010 | -0.0737 | No |
| 54 | Acox1 | 6384 | -0.017 | -0.0806 | No |
| 55 | Gabarapl1 | 6537 | -0.028 | -0.0921 | No |
| 56 | Shmt2 | 6772 | -0.044 | -0.1098 | No |
| 57 | Ninj1 | 6820 | -0.047 | -0.1121 | No |
| 58 | Pcx | 6862 | -0.050 | -0.1138 | No |
| 59 | Casp6 | 6942 | -0.055 | -0.1184 | No |
| 60 | Tmem176b | 6951 | -0.056 | -0.1172 | No |
| 61 | Aqp9 | 7077 | -0.064 | -0.1254 | No |
| 62 | Cat | 7096 | -0.065 | -0.1247 | No |
| 63 | Pgrmc1 | 7149 | -0.068 | -0.1267 | No |
| 64 | Gss | 7278 | -0.076 | -0.1347 | No |
| 65 | Cyfip2 | 7304 | -0.077 | -0.1342 | No |
| 66 | Asl | 7396 | -0.083 | -0.1389 | No |
| 67 | Ets2 | 7436 | -0.085 | -0.1393 | No |
| 68 | Angptl3 | 7640 | -0.101 | -0.1526 | No |
| 69 | Hgfac | 7652 | -0.102 | -0.1502 | No |
| 70 | Nmt1 | 7681 | -0.103 | -0.1491 | No |
| 71 | Slc35d1 | 7701 | -0.104 | -0.1472 | No |
| 72 | Nfs1 | 7708 | -0.105 | -0.1443 | No |
| 73 | Pink1 | 7836 | -0.114 | -0.1509 | No |
| 74 | Ssr3 | 8583 | -0.163 | -0.2065 | No |
| 75 | Aldh9a1 | 8837 | -0.183 | -0.2212 | No |
| 76 | Gcnt2 | 8888 | -0.187 | -0.2192 | No |
| 77 | Ptges | 9087 | -0.202 | -0.2287 | No |
| 78 | Il1r1 | 9288 | -0.216 | -0.2380 | Yes |
| 79 | Adh5 | 9306 | -0.218 | -0.2322 | Yes |
| 80 | Comt | 9316 | -0.218 | -0.2258 | Yes |
| 81 | Npc1 | 9320 | -0.219 | -0.2189 | Yes |
| 82 | Spint2 | 9484 | -0.233 | -0.2246 | Yes |
| 83 | Ap4b1 | 9658 | -0.249 | -0.2306 | Yes |
| 84 | Dcxr | 9710 | -0.253 | -0.2265 | Yes |
| 85 | Alas1 | 9747 | -0.256 | -0.2210 | Yes |
| 86 | Por | 9758 | -0.257 | -0.2135 | Yes |
| 87 | Tkfc | 9789 | -0.260 | -0.2074 | Yes |
| 88 | Gsr | 9799 | -0.260 | -0.1996 | Yes |
| 89 | Dhrs7 | 10078 | -0.285 | -0.2130 | Yes |
| 90 | Kynu | 10086 | -0.285 | -0.2042 | Yes |
| 91 | Gch1 | 10087 | -0.285 | -0.1949 | Yes |
| 92 | Cyp27a1 | 10173 | -0.293 | -0.1922 | Yes |
| 93 | Apoe | 10182 | -0.294 | -0.1833 | Yes |
| 94 | Irf8 | 10249 | -0.301 | -0.1788 | Yes |
| 95 | Gstm4 | 10478 | -0.322 | -0.1869 | Yes |
| 96 | Slc46a3 | 10501 | -0.326 | -0.1780 | Yes |
| 97 | Ech1 | 10718 | -0.348 | -0.1843 | Yes |
| 98 | Etfdh | 10728 | -0.349 | -0.1736 | Yes |
| 99 | Tpst1 | 10845 | -0.364 | -0.1712 | Yes |
| 100 | Ugdh | 10931 | -0.376 | -0.1658 | Yes |
| 101 | Entpd5 | 10988 | -0.386 | -0.1578 | Yes |
| 102 | Ppard | 11123 | -0.404 | -0.1555 | Yes |
| 103 | Xdh | 11173 | -0.414 | -0.1459 | Yes |
| 104 | Psmb10 | 11240 | -0.424 | -0.1375 | Yes |
| 105 | Cbr1 | 11274 | -0.429 | -0.1261 | Yes |
| 106 | Pmm1 | 11425 | -0.456 | -0.1234 | Yes |
| 107 | Retsat | 11487 | -0.466 | -0.1132 | Yes |
| 108 | Gstt2 | 11603 | -0.488 | -0.1066 | Yes |
| 109 | Tnfrsf1a | 11609 | -0.489 | -0.0910 | Yes |
| 110 | Sertad1 | 11643 | -0.496 | -0.0775 | Yes |
| 111 | Acp2 | 11666 | -0.501 | -0.0629 | Yes |
| 112 | Tmbim6 | 11886 | -0.553 | -0.0627 | Yes |
| 113 | Idh1 | 11926 | -0.566 | -0.0473 | Yes |
| 114 | Acox3 | 12014 | -0.599 | -0.0348 | Yes |
| 115 | Car2 | 12048 | -0.612 | -0.0175 | Yes |
| 116 | Esr1 | 12078 | -0.629 | 0.0007 | Yes |
| 117 | Atp2a2 | 12172 | -0.687 | 0.0156 | Yes |