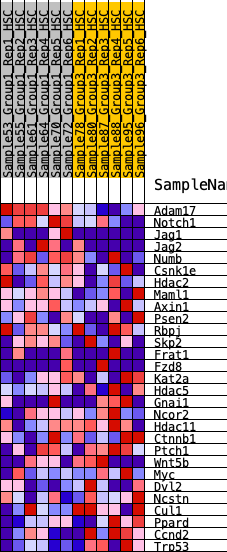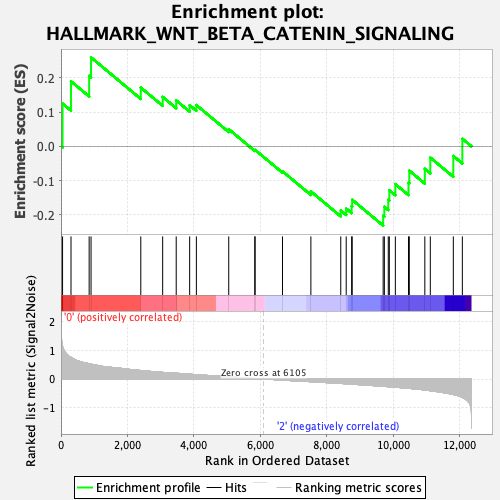
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.25996307 |
| Normalized Enrichment Score (NES) | 0.91068137 |
| Nominal p-value | 0.6044625 |
| FDR q-value | 0.9075559 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adam17 | 47 | 1.133 | 0.1251 | Yes |
| 2 | Notch1 | 301 | 0.752 | 0.1901 | Yes |
| 3 | Jag1 | 848 | 0.529 | 0.2060 | Yes |
| 4 | Jag2 | 905 | 0.514 | 0.2600 | Yes |
| 5 | Numb | 2403 | 0.293 | 0.1719 | No |
| 6 | Csnk1e | 3062 | 0.228 | 0.1446 | No |
| 7 | Hdac2 | 3470 | 0.199 | 0.1343 | No |
| 8 | Maml1 | 3875 | 0.165 | 0.1203 | No |
| 9 | Axin1 | 4077 | 0.148 | 0.1208 | No |
| 10 | Psen2 | 5052 | 0.073 | 0.0502 | No |
| 11 | Rbpj | 5837 | 0.017 | -0.0114 | No |
| 12 | Skp2 | 5846 | 0.017 | -0.0102 | No |
| 13 | Frat1 | 6671 | -0.037 | -0.0727 | No |
| 14 | Fzd8 | 7526 | -0.092 | -0.1316 | No |
| 15 | Kat2a | 8426 | -0.153 | -0.1871 | No |
| 16 | Hdac5 | 8590 | -0.163 | -0.1817 | No |
| 17 | Gnai1 | 8754 | -0.177 | -0.1748 | No |
| 18 | Ncor2 | 8770 | -0.178 | -0.1558 | No |
| 19 | Hdac11 | 9703 | -0.252 | -0.2027 | No |
| 20 | Ctnnb1 | 9739 | -0.255 | -0.1765 | No |
| 21 | Ptch1 | 9857 | -0.264 | -0.1559 | No |
| 22 | Wnt5b | 9889 | -0.267 | -0.1280 | No |
| 23 | Myc | 10070 | -0.284 | -0.1102 | No |
| 24 | Dvl2 | 10466 | -0.320 | -0.1058 | No |
| 25 | Ncstn | 10490 | -0.324 | -0.0708 | No |
| 26 | Cul1 | 10958 | -0.380 | -0.0654 | No |
| 27 | Ppard | 11123 | -0.404 | -0.0327 | No |
| 28 | Ccnd2 | 11816 | -0.535 | -0.0280 | No |
| 29 | Trp53 | 12088 | -0.635 | 0.0223 | No |