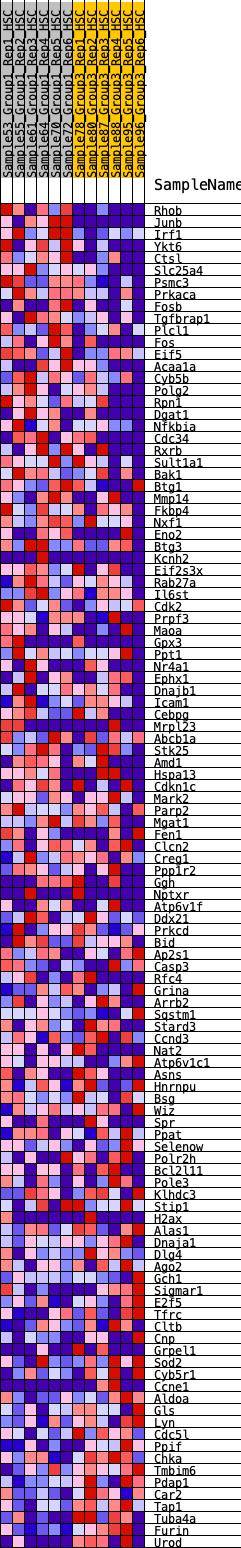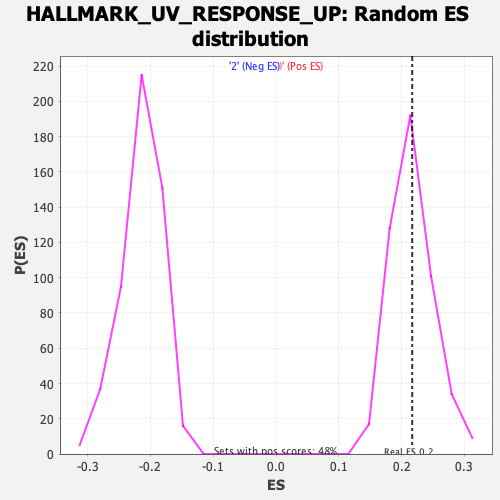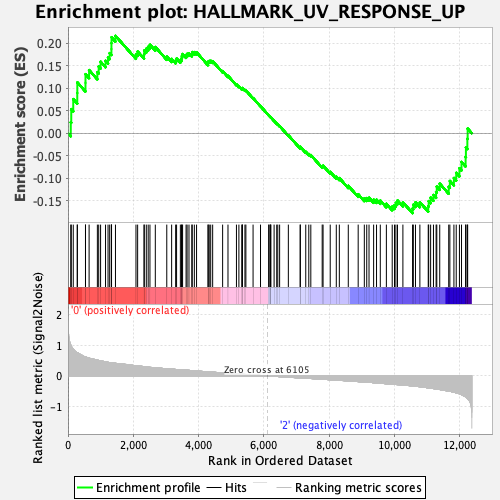
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.21692285 |
| Normalized Enrichment Score (NES) | 1.0018638 |
| Nominal p-value | 0.47401246 |
| FDR q-value | 0.88835907 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rhob | 85 | 1.009 | 0.0244 | Yes |
| 2 | Junb | 100 | 0.977 | 0.0537 | Yes |
| 3 | Irf1 | 161 | 0.874 | 0.0760 | Yes |
| 4 | Ykt6 | 284 | 0.760 | 0.0897 | Yes |
| 5 | Ctsl | 286 | 0.759 | 0.1132 | Yes |
| 6 | Slc25a4 | 535 | 0.619 | 0.1123 | Yes |
| 7 | Psmc3 | 536 | 0.619 | 0.1315 | Yes |
| 8 | Prkaca | 646 | 0.580 | 0.1407 | Yes |
| 9 | Fosb | 899 | 0.515 | 0.1361 | Yes |
| 10 | Tgfbrap1 | 940 | 0.505 | 0.1486 | Yes |
| 11 | Plcl1 | 998 | 0.493 | 0.1593 | Yes |
| 12 | Fos | 1149 | 0.460 | 0.1613 | Yes |
| 13 | Eif5 | 1226 | 0.444 | 0.1690 | Yes |
| 14 | Acaa1a | 1276 | 0.433 | 0.1784 | Yes |
| 15 | Cyb5b | 1329 | 0.427 | 0.1875 | Yes |
| 16 | Polg2 | 1330 | 0.427 | 0.2007 | Yes |
| 17 | Rpn1 | 1333 | 0.426 | 0.2138 | Yes |
| 18 | Dgat1 | 1453 | 0.412 | 0.2169 | Yes |
| 19 | Nfkbia | 2081 | 0.329 | 0.1760 | No |
| 20 | Cdc34 | 2132 | 0.324 | 0.1820 | No |
| 21 | Rxrb | 2328 | 0.303 | 0.1755 | No |
| 22 | Sult1a1 | 2334 | 0.302 | 0.1845 | No |
| 23 | Bak1 | 2397 | 0.294 | 0.1886 | No |
| 24 | Btg1 | 2455 | 0.286 | 0.1928 | No |
| 25 | Mmp14 | 2509 | 0.279 | 0.1972 | No |
| 26 | Fkbp4 | 2673 | 0.265 | 0.1921 | No |
| 27 | Nxf1 | 3024 | 0.231 | 0.1707 | No |
| 28 | Eno2 | 3173 | 0.221 | 0.1656 | No |
| 29 | Btg3 | 3296 | 0.210 | 0.1621 | No |
| 30 | Kcnh2 | 3324 | 0.209 | 0.1664 | No |
| 31 | Eif2s3x | 3440 | 0.202 | 0.1633 | No |
| 32 | Rab27a | 3475 | 0.199 | 0.1667 | No |
| 33 | Il6st | 3487 | 0.198 | 0.1720 | No |
| 34 | Cdk2 | 3508 | 0.196 | 0.1765 | No |
| 35 | Prpf3 | 3615 | 0.188 | 0.1737 | No |
| 36 | Maoa | 3646 | 0.186 | 0.1770 | No |
| 37 | Gpx3 | 3704 | 0.180 | 0.1779 | No |
| 38 | Ppt1 | 3792 | 0.173 | 0.1762 | No |
| 39 | Nr4a1 | 3802 | 0.172 | 0.1808 | No |
| 40 | Ephx1 | 3868 | 0.165 | 0.1807 | No |
| 41 | Dnajb1 | 3936 | 0.160 | 0.1802 | No |
| 42 | Icam1 | 4289 | 0.132 | 0.1556 | No |
| 43 | Cebpg | 4290 | 0.132 | 0.1597 | No |
| 44 | Mrpl23 | 4323 | 0.129 | 0.1611 | No |
| 45 | Abcb1a | 4369 | 0.127 | 0.1614 | No |
| 46 | Stk25 | 4429 | 0.122 | 0.1603 | No |
| 47 | Amd1 | 4734 | 0.096 | 0.1385 | No |
| 48 | Hspa13 | 4898 | 0.084 | 0.1278 | No |
| 49 | Cdkn1c | 5157 | 0.065 | 0.1088 | No |
| 50 | Mark2 | 5232 | 0.059 | 0.1046 | No |
| 51 | Parp2 | 5321 | 0.053 | 0.0991 | No |
| 52 | Mgat1 | 5338 | 0.052 | 0.0994 | No |
| 53 | Fen1 | 5341 | 0.052 | 0.1008 | No |
| 54 | Clcn2 | 5417 | 0.047 | 0.0962 | No |
| 55 | Creg1 | 5446 | 0.045 | 0.0953 | No |
| 56 | Ppp1r2 | 5666 | 0.030 | 0.0784 | No |
| 57 | Ggh | 5894 | 0.013 | 0.0602 | No |
| 58 | Nptxr | 6143 | -0.000 | 0.0400 | No |
| 59 | Atp6v1f | 6158 | -0.001 | 0.0389 | No |
| 60 | Ddx21 | 6195 | -0.003 | 0.0361 | No |
| 61 | Prkcd | 6204 | -0.004 | 0.0355 | No |
| 62 | Bid | 6209 | -0.004 | 0.0354 | No |
| 63 | Ap2s1 | 6310 | -0.011 | 0.0275 | No |
| 64 | Casp3 | 6390 | -0.017 | 0.0216 | No |
| 65 | Rfc4 | 6423 | -0.020 | 0.0196 | No |
| 66 | Grina | 6479 | -0.024 | 0.0159 | No |
| 67 | Arrb2 | 6746 | -0.042 | -0.0045 | No |
| 68 | Sqstm1 | 7109 | -0.066 | -0.0320 | No |
| 69 | Stard3 | 7112 | -0.066 | -0.0301 | No |
| 70 | Ccnd3 | 7279 | -0.076 | -0.0413 | No |
| 71 | Nat2 | 7372 | -0.082 | -0.0463 | No |
| 72 | Atp6v1c1 | 7434 | -0.085 | -0.0486 | No |
| 73 | Asns | 7782 | -0.110 | -0.0735 | No |
| 74 | Hnrnpu | 7809 | -0.112 | -0.0722 | No |
| 75 | Bsg | 8028 | -0.125 | -0.0861 | No |
| 76 | Wiz | 8214 | -0.139 | -0.0969 | No |
| 77 | Spr | 8307 | -0.145 | -0.0998 | No |
| 78 | Ppat | 8581 | -0.162 | -0.1171 | No |
| 79 | Selenow | 8884 | -0.186 | -0.1359 | No |
| 80 | Polr2h | 9072 | -0.201 | -0.1449 | No |
| 81 | Bcl2l11 | 9146 | -0.207 | -0.1445 | No |
| 82 | Pole3 | 9216 | -0.210 | -0.1435 | No |
| 83 | Klhdc3 | 9357 | -0.223 | -0.1481 | No |
| 84 | Stip1 | 9446 | -0.230 | -0.1481 | No |
| 85 | H2ax | 9561 | -0.240 | -0.1499 | No |
| 86 | Alas1 | 9747 | -0.256 | -0.1570 | No |
| 87 | Dnaja1 | 9926 | -0.271 | -0.1631 | No |
| 88 | Dlg4 | 9996 | -0.277 | -0.1601 | No |
| 89 | Ago2 | 10039 | -0.281 | -0.1548 | No |
| 90 | Gch1 | 10087 | -0.285 | -0.1498 | No |
| 91 | Sigmar1 | 10253 | -0.301 | -0.1539 | No |
| 92 | E2f5 | 10549 | -0.330 | -0.1677 | No |
| 93 | Tfrc | 10572 | -0.333 | -0.1591 | No |
| 94 | Cltb | 10638 | -0.340 | -0.1538 | No |
| 95 | Cnp | 10773 | -0.354 | -0.1538 | No |
| 96 | Grpel1 | 11029 | -0.391 | -0.1624 | No |
| 97 | Sod2 | 11043 | -0.392 | -0.1512 | No |
| 98 | Cyb5r1 | 11102 | -0.402 | -0.1435 | No |
| 99 | Ccne1 | 11189 | -0.416 | -0.1375 | No |
| 100 | Aldoa | 11270 | -0.429 | -0.1307 | No |
| 101 | Gls | 11291 | -0.431 | -0.1190 | No |
| 102 | Lyn | 11381 | -0.446 | -0.1124 | No |
| 103 | Cdc5l | 11656 | -0.498 | -0.1192 | No |
| 104 | Ppif | 11691 | -0.506 | -0.1062 | No |
| 105 | Chka | 11815 | -0.534 | -0.0997 | No |
| 106 | Tmbim6 | 11886 | -0.553 | -0.0882 | No |
| 107 | Pdap1 | 11984 | -0.587 | -0.0778 | No |
| 108 | Car2 | 12048 | -0.612 | -0.0639 | No |
| 109 | Tap1 | 12173 | -0.687 | -0.0526 | No |
| 110 | Tuba4a | 12183 | -0.693 | -0.0318 | No |
| 111 | Furin | 12227 | -0.740 | -0.0123 | No |
| 112 | Urod | 12236 | -0.749 | 0.0104 | No |