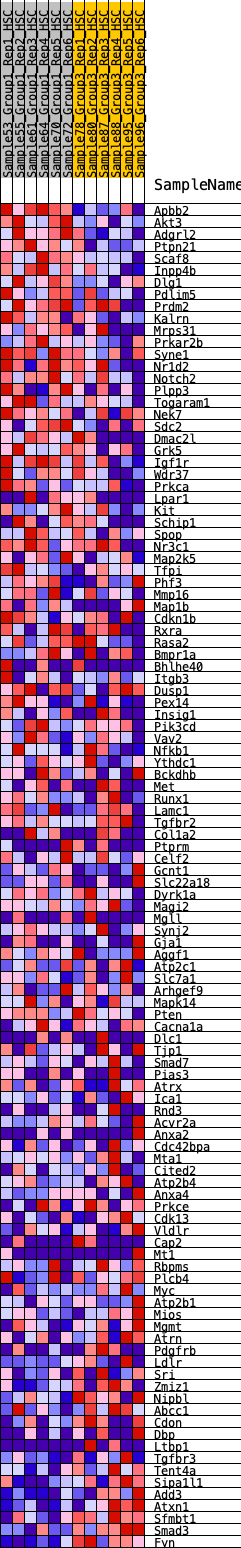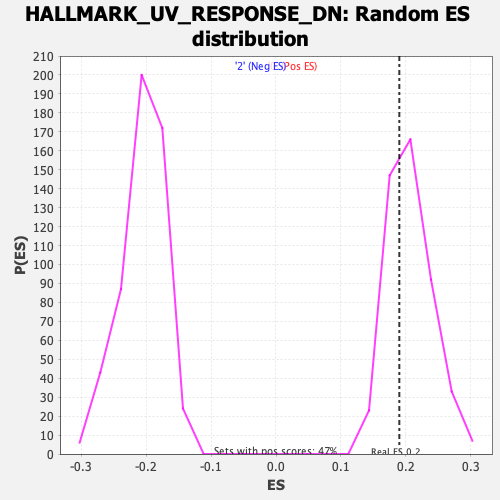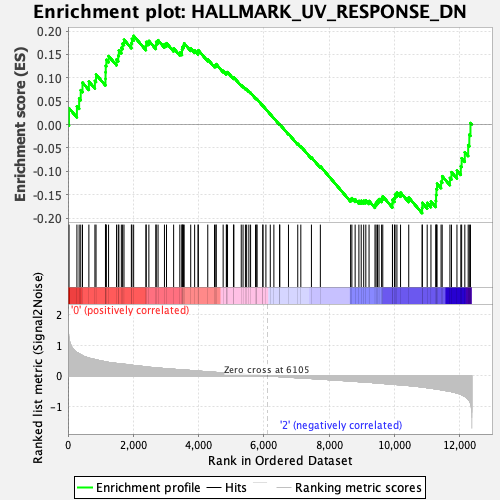
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.1897665 |
| Normalized Enrichment Score (NES) | 0.92218107 |
| Nominal p-value | 0.6517094 |
| FDR q-value | 0.9383403 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Apbb2 | 31 | 1.212 | 0.0350 | Yes |
| 2 | Akt3 | 273 | 0.767 | 0.0390 | Yes |
| 3 | Adgrl2 | 341 | 0.725 | 0.0560 | Yes |
| 4 | Ptpn21 | 390 | 0.697 | 0.0736 | Yes |
| 5 | Scaf8 | 444 | 0.664 | 0.0898 | Yes |
| 6 | Inpp4b | 639 | 0.582 | 0.0920 | Yes |
| 7 | Dlg1 | 824 | 0.537 | 0.0936 | Yes |
| 8 | Pdlim5 | 858 | 0.526 | 0.1072 | Yes |
| 9 | Prdm2 | 1147 | 0.460 | 0.0979 | Yes |
| 10 | Kalrn | 1150 | 0.460 | 0.1120 | Yes |
| 11 | Mrps31 | 1156 | 0.458 | 0.1258 | Yes |
| 12 | Prkar2b | 1174 | 0.456 | 0.1385 | Yes |
| 13 | Syne1 | 1242 | 0.440 | 0.1466 | Yes |
| 14 | Nr1d2 | 1486 | 0.406 | 0.1393 | Yes |
| 15 | Notch2 | 1538 | 0.401 | 0.1476 | Yes |
| 16 | Plpp3 | 1557 | 0.398 | 0.1584 | Yes |
| 17 | Togaram1 | 1633 | 0.391 | 0.1644 | Yes |
| 18 | Nek7 | 1672 | 0.385 | 0.1732 | Yes |
| 19 | Sdc2 | 1714 | 0.380 | 0.1816 | Yes |
| 20 | Dmac2l | 1940 | 0.350 | 0.1740 | Yes |
| 21 | Grk5 | 1957 | 0.348 | 0.1835 | Yes |
| 22 | Igf1r | 2010 | 0.340 | 0.1898 | Yes |
| 23 | Wdr37 | 2386 | 0.296 | 0.1683 | No |
| 24 | Prkca | 2394 | 0.294 | 0.1768 | No |
| 25 | Lpar1 | 2476 | 0.283 | 0.1790 | No |
| 26 | Kit | 2690 | 0.263 | 0.1698 | No |
| 27 | Schip1 | 2700 | 0.262 | 0.1771 | No |
| 28 | Spop | 2757 | 0.258 | 0.1805 | No |
| 29 | Nr3c1 | 2953 | 0.238 | 0.1720 | No |
| 30 | Map2k5 | 3016 | 0.233 | 0.1741 | No |
| 31 | Tfpi | 3234 | 0.216 | 0.1631 | No |
| 32 | Phf3 | 3424 | 0.203 | 0.1540 | No |
| 33 | Mmp16 | 3490 | 0.198 | 0.1548 | No |
| 34 | Map1b | 3491 | 0.198 | 0.1609 | No |
| 35 | Cdkn1b | 3503 | 0.197 | 0.1661 | No |
| 36 | Rxra | 3537 | 0.194 | 0.1694 | No |
| 37 | Rasa2 | 3556 | 0.193 | 0.1738 | No |
| 38 | Bmpr1a | 3757 | 0.176 | 0.1630 | No |
| 39 | Bhlhe40 | 3877 | 0.165 | 0.1583 | No |
| 40 | Itgb3 | 3981 | 0.157 | 0.1548 | No |
| 41 | Dusp1 | 3990 | 0.156 | 0.1589 | No |
| 42 | Pex14 | 4279 | 0.133 | 0.1395 | No |
| 43 | Insig1 | 4485 | 0.117 | 0.1264 | No |
| 44 | Pik3cd | 4528 | 0.113 | 0.1265 | No |
| 45 | Vav2 | 4536 | 0.112 | 0.1294 | No |
| 46 | Nfkb1 | 4750 | 0.095 | 0.1149 | No |
| 47 | Ythdc1 | 4847 | 0.088 | 0.1098 | No |
| 48 | Bckdhb | 4855 | 0.087 | 0.1119 | No |
| 49 | Met | 4884 | 0.085 | 0.1123 | No |
| 50 | Runx1 | 5066 | 0.071 | 0.0997 | No |
| 51 | Lamc1 | 5079 | 0.070 | 0.1009 | No |
| 52 | Tgfbr2 | 5304 | 0.055 | 0.0843 | No |
| 53 | Col1a2 | 5359 | 0.051 | 0.0815 | No |
| 54 | Ptprm | 5432 | 0.046 | 0.0770 | No |
| 55 | Celf2 | 5469 | 0.043 | 0.0754 | No |
| 56 | Gcnt1 | 5537 | 0.039 | 0.0712 | No |
| 57 | Slc22a18 | 5590 | 0.035 | 0.0680 | No |
| 58 | Dyrk1a | 5741 | 0.025 | 0.0565 | No |
| 59 | Magi2 | 5782 | 0.022 | 0.0539 | No |
| 60 | Mgll | 5783 | 0.022 | 0.0546 | No |
| 61 | Synj2 | 5966 | 0.009 | 0.0400 | No |
| 62 | Gja1 | 5967 | 0.009 | 0.0403 | No |
| 63 | Aggf1 | 6059 | 0.002 | 0.0329 | No |
| 64 | Atp2c1 | 6193 | -0.003 | 0.0222 | No |
| 65 | Slc7a1 | 6303 | -0.011 | 0.0136 | No |
| 66 | Arhgef9 | 6477 | -0.024 | 0.0003 | No |
| 67 | Mapk14 | 6484 | -0.025 | 0.0005 | No |
| 68 | Pten | 6750 | -0.042 | -0.0198 | No |
| 69 | Cacna1a | 7034 | -0.061 | -0.0410 | No |
| 70 | Dlc1 | 7128 | -0.067 | -0.0465 | No |
| 71 | Tjp1 | 7453 | -0.086 | -0.0703 | No |
| 72 | Smad7 | 7726 | -0.106 | -0.0892 | No |
| 73 | Pias3 | 8652 | -0.168 | -0.1595 | No |
| 74 | Atrx | 8691 | -0.172 | -0.1573 | No |
| 75 | Ica1 | 8791 | -0.179 | -0.1598 | No |
| 76 | Rnd3 | 8906 | -0.189 | -0.1633 | No |
| 77 | Acvr2a | 8977 | -0.194 | -0.1630 | No |
| 78 | Anxa2 | 9053 | -0.199 | -0.1630 | No |
| 79 | Cdc42bpa | 9118 | -0.205 | -0.1619 | No |
| 80 | Mta1 | 9217 | -0.210 | -0.1634 | No |
| 81 | Cited2 | 9399 | -0.227 | -0.1711 | No |
| 82 | Atp2b4 | 9433 | -0.229 | -0.1667 | No |
| 83 | Anxa4 | 9476 | -0.233 | -0.1630 | No |
| 84 | Prkce | 9522 | -0.237 | -0.1593 | No |
| 85 | Cdk13 | 9601 | -0.243 | -0.1582 | No |
| 86 | Vldlr | 9637 | -0.247 | -0.1534 | No |
| 87 | Cap2 | 9933 | -0.272 | -0.1691 | No |
| 88 | Mt1 | 9935 | -0.272 | -0.1607 | No |
| 89 | Rbpms | 9995 | -0.277 | -0.1570 | No |
| 90 | Plcb4 | 10013 | -0.278 | -0.1498 | No |
| 91 | Myc | 10070 | -0.284 | -0.1455 | No |
| 92 | Atp2b1 | 10181 | -0.294 | -0.1454 | No |
| 93 | Mios | 10432 | -0.317 | -0.1560 | No |
| 94 | Mgmt | 10842 | -0.363 | -0.1782 | No |
| 95 | Atrn | 10848 | -0.364 | -0.1673 | No |
| 96 | Pdgfrb | 10997 | -0.387 | -0.1675 | No |
| 97 | Ldlr | 11112 | -0.403 | -0.1643 | No |
| 98 | Sri | 11261 | -0.427 | -0.1632 | No |
| 99 | Zmiz1 | 11269 | -0.428 | -0.1505 | No |
| 100 | Nipbl | 11280 | -0.430 | -0.1380 | No |
| 101 | Abcc1 | 11299 | -0.433 | -0.1261 | No |
| 102 | Cdon | 11422 | -0.455 | -0.1220 | No |
| 103 | Dbp | 11456 | -0.462 | -0.1104 | No |
| 104 | Ltbp1 | 11693 | -0.506 | -0.1140 | No |
| 105 | Tgfbr3 | 11740 | -0.517 | -0.1017 | No |
| 106 | Tent4a | 11908 | -0.561 | -0.0980 | No |
| 107 | Sipa1l1 | 12027 | -0.604 | -0.0890 | No |
| 108 | Add3 | 12052 | -0.614 | -0.0720 | No |
| 109 | Atxn1 | 12149 | -0.669 | -0.0591 | No |
| 110 | Sfmbt1 | 12252 | -0.765 | -0.0438 | No |
| 111 | Smad3 | 12287 | -0.813 | -0.0214 | No |
| 112 | Fyn | 12322 | -0.890 | 0.0033 | No |