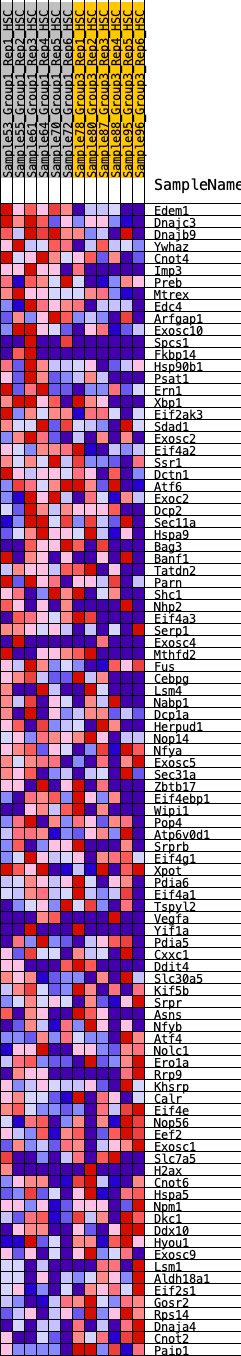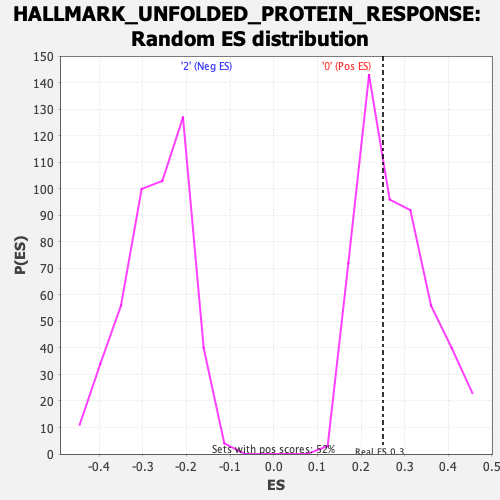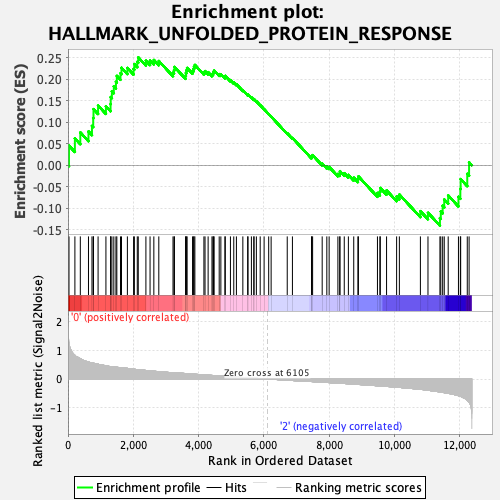
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.25032255 |
| Normalized Enrichment Score (NES) | 0.9032125 |
| Nominal p-value | 0.552381 |
| FDR q-value | 0.81970704 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Edem1 | 30 | 1.238 | 0.0455 | Yes |
| 2 | Dnajc3 | 211 | 0.816 | 0.0624 | Yes |
| 3 | Dnajb9 | 377 | 0.702 | 0.0762 | Yes |
| 4 | Ywhaz | 627 | 0.584 | 0.0785 | Yes |
| 5 | Cnot4 | 734 | 0.559 | 0.0915 | Yes |
| 6 | Imp3 | 772 | 0.549 | 0.1097 | Yes |
| 7 | Preb | 781 | 0.547 | 0.1303 | Yes |
| 8 | Mtrex | 921 | 0.510 | 0.1387 | Yes |
| 9 | Edc4 | 1161 | 0.458 | 0.1369 | Yes |
| 10 | Arfgap1 | 1303 | 0.429 | 0.1420 | Yes |
| 11 | Exosc10 | 1308 | 0.428 | 0.1583 | Yes |
| 12 | Spcs1 | 1344 | 0.423 | 0.1718 | Yes |
| 13 | Fkbp14 | 1403 | 0.416 | 0.1832 | Yes |
| 14 | Hsp90b1 | 1468 | 0.410 | 0.1938 | Yes |
| 15 | Psat1 | 1495 | 0.406 | 0.2074 | Yes |
| 16 | Ern1 | 1607 | 0.393 | 0.2136 | Yes |
| 17 | Xbp1 | 1638 | 0.390 | 0.2262 | Yes |
| 18 | Eif2ak3 | 1818 | 0.367 | 0.2259 | Yes |
| 19 | Sdad1 | 2008 | 0.341 | 0.2236 | Yes |
| 20 | Exosc2 | 2032 | 0.337 | 0.2348 | Yes |
| 21 | Eif4a2 | 2121 | 0.325 | 0.2402 | Yes |
| 22 | Ssr1 | 2151 | 0.322 | 0.2503 | Yes |
| 23 | Dctn1 | 2385 | 0.296 | 0.2428 | No |
| 24 | Atf6 | 2512 | 0.279 | 0.2433 | No |
| 25 | Exoc2 | 2628 | 0.269 | 0.2443 | No |
| 26 | Dcp2 | 2780 | 0.256 | 0.2419 | No |
| 27 | Sec11a | 3215 | 0.218 | 0.2150 | No |
| 28 | Hspa9 | 3246 | 0.215 | 0.2209 | No |
| 29 | Bag3 | 3259 | 0.213 | 0.2282 | No |
| 30 | Banf1 | 3599 | 0.189 | 0.2078 | No |
| 31 | Tatdn2 | 3610 | 0.188 | 0.2143 | No |
| 32 | Parn | 3625 | 0.187 | 0.2204 | No |
| 33 | Shc1 | 3647 | 0.186 | 0.2259 | No |
| 34 | Nhp2 | 3813 | 0.171 | 0.2191 | No |
| 35 | Eif4a3 | 3839 | 0.168 | 0.2235 | No |
| 36 | Serp1 | 3858 | 0.167 | 0.2285 | No |
| 37 | Exosc4 | 3884 | 0.164 | 0.2329 | No |
| 38 | Mthfd2 | 4161 | 0.141 | 0.2158 | No |
| 39 | Fus | 4195 | 0.139 | 0.2185 | No |
| 40 | Cebpg | 4290 | 0.132 | 0.2160 | No |
| 41 | Lsm4 | 4408 | 0.124 | 0.2112 | No |
| 42 | Nabp1 | 4438 | 0.121 | 0.2136 | No |
| 43 | Dcp1a | 4454 | 0.119 | 0.2170 | No |
| 44 | Herpud1 | 4474 | 0.118 | 0.2200 | No |
| 45 | Nop14 | 4629 | 0.105 | 0.2115 | No |
| 46 | Nfya | 4680 | 0.100 | 0.2113 | No |
| 47 | Exosc5 | 4804 | 0.091 | 0.2048 | No |
| 48 | Sec31a | 4815 | 0.091 | 0.2075 | No |
| 49 | Zbtb17 | 4973 | 0.079 | 0.1977 | No |
| 50 | Eif4ebp1 | 5075 | 0.071 | 0.1922 | No |
| 51 | Wipi1 | 5156 | 0.065 | 0.1882 | No |
| 52 | Pop4 | 5353 | 0.051 | 0.1742 | No |
| 53 | Atp6v0d1 | 5501 | 0.041 | 0.1638 | No |
| 54 | Srprb | 5511 | 0.040 | 0.1646 | No |
| 55 | Eif4g1 | 5615 | 0.033 | 0.1575 | No |
| 56 | Xpot | 5689 | 0.028 | 0.1527 | No |
| 57 | Pdia6 | 5695 | 0.028 | 0.1533 | No |
| 58 | Eif4a1 | 5767 | 0.023 | 0.1484 | No |
| 59 | Tspyl2 | 5884 | 0.014 | 0.1395 | No |
| 60 | Vegfa | 6004 | 0.006 | 0.1301 | No |
| 61 | Yif1a | 6144 | -0.000 | 0.1187 | No |
| 62 | Pdia5 | 6219 | -0.005 | 0.1129 | No |
| 63 | Cxxc1 | 6712 | -0.040 | 0.0743 | No |
| 64 | Ddit4 | 6871 | -0.050 | 0.0634 | No |
| 65 | Slc30a5 | 7456 | -0.087 | 0.0192 | No |
| 66 | Kif5b | 7465 | -0.087 | 0.0219 | No |
| 67 | Srpr | 7489 | -0.089 | 0.0235 | No |
| 68 | Asns | 7782 | -0.110 | 0.0039 | No |
| 69 | Nfyb | 7924 | -0.118 | -0.0030 | No |
| 70 | Atf4 | 7995 | -0.123 | -0.0039 | No |
| 71 | Nolc1 | 8263 | -0.143 | -0.0202 | No |
| 72 | Ero1a | 8320 | -0.146 | -0.0191 | No |
| 73 | Rrp9 | 8326 | -0.147 | -0.0138 | No |
| 74 | Khsrp | 8458 | -0.155 | -0.0185 | No |
| 75 | Calr | 8584 | -0.163 | -0.0224 | No |
| 76 | Eif4e | 8748 | -0.176 | -0.0288 | No |
| 77 | Nop56 | 8876 | -0.186 | -0.0320 | No |
| 78 | Eef2 | 8891 | -0.188 | -0.0259 | No |
| 79 | Exosc1 | 9474 | -0.232 | -0.0643 | No |
| 80 | Slc7a5 | 9549 | -0.238 | -0.0611 | No |
| 81 | H2ax | 9561 | -0.240 | -0.0527 | No |
| 82 | Cnot6 | 9753 | -0.256 | -0.0584 | No |
| 83 | Hspa5 | 10061 | -0.284 | -0.0724 | No |
| 84 | Npm1 | 10145 | -0.290 | -0.0679 | No |
| 85 | Dkc1 | 10789 | -0.356 | -0.1065 | No |
| 86 | Ddx10 | 11021 | -0.390 | -0.1103 | No |
| 87 | Hyou1 | 11387 | -0.448 | -0.1227 | No |
| 88 | Exosc9 | 11414 | -0.453 | -0.1072 | No |
| 89 | Lsm1 | 11471 | -0.464 | -0.0939 | No |
| 90 | Aldh18a1 | 11519 | -0.471 | -0.0794 | No |
| 91 | Eif2s1 | 11640 | -0.496 | -0.0700 | No |
| 92 | Gosr2 | 11954 | -0.576 | -0.0732 | No |
| 93 | Rps14 | 12012 | -0.598 | -0.0547 | No |
| 94 | Dnaja4 | 12022 | -0.603 | -0.0321 | No |
| 95 | Cnot2 | 12224 | -0.738 | -0.0199 | No |
| 96 | Paip1 | 12280 | -0.805 | 0.0068 | No |