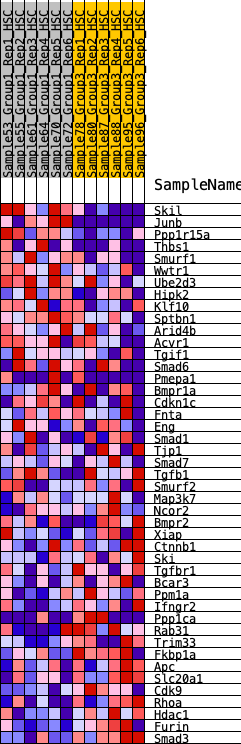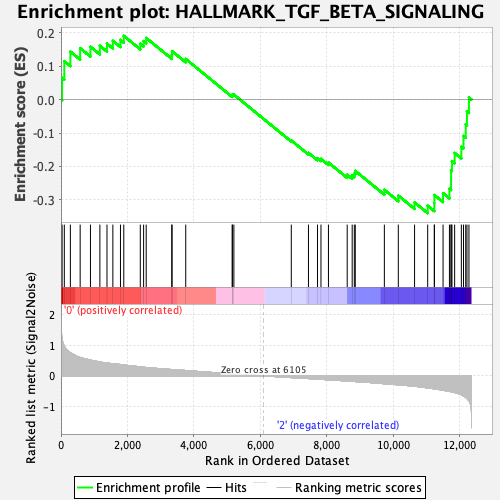
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.3396618 |
| Normalized Enrichment Score (NES) | -1.2913568 |
| Nominal p-value | 0.10788382 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.838 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Skil | 33 | 1.193 | 0.0650 | No |
| 2 | Junb | 100 | 0.977 | 0.1150 | No |
| 3 | Ppp1r15a | 282 | 0.760 | 0.1434 | No |
| 4 | Thbs1 | 577 | 0.605 | 0.1538 | No |
| 5 | Smurf1 | 887 | 0.519 | 0.1581 | No |
| 6 | Wwtr1 | 1171 | 0.456 | 0.1610 | No |
| 7 | Ube2d3 | 1385 | 0.419 | 0.1674 | No |
| 8 | Hipk2 | 1562 | 0.397 | 0.1757 | No |
| 9 | Klf10 | 1791 | 0.373 | 0.1783 | No |
| 10 | Sptbn1 | 1891 | 0.358 | 0.1906 | No |
| 11 | Arid4b | 2388 | 0.295 | 0.1670 | No |
| 12 | Acvr1 | 2488 | 0.282 | 0.1749 | No |
| 13 | Tgif1 | 2566 | 0.275 | 0.1843 | No |
| 14 | Smad6 | 3336 | 0.209 | 0.1337 | No |
| 15 | Pmepa1 | 3345 | 0.208 | 0.1448 | No |
| 16 | Bmpr1a | 3757 | 0.176 | 0.1214 | No |
| 17 | Cdkn1c | 5157 | 0.065 | 0.0115 | No |
| 18 | Fnta | 5161 | 0.064 | 0.0149 | No |
| 19 | Eng | 5207 | 0.061 | 0.0147 | No |
| 20 | Smad1 | 6936 | -0.055 | -0.1224 | No |
| 21 | Tjp1 | 7453 | -0.086 | -0.1594 | No |
| 22 | Smad7 | 7726 | -0.106 | -0.1755 | No |
| 23 | Tgfb1 | 7830 | -0.113 | -0.1774 | No |
| 24 | Smurf2 | 8055 | -0.127 | -0.1884 | No |
| 25 | Map3k7 | 8620 | -0.166 | -0.2248 | No |
| 26 | Ncor2 | 8770 | -0.178 | -0.2268 | No |
| 27 | Bmpr2 | 8834 | -0.183 | -0.2215 | No |
| 28 | Xiap | 8863 | -0.185 | -0.2133 | No |
| 29 | Ctnnb1 | 9739 | -0.255 | -0.2699 | No |
| 30 | Ski | 10159 | -0.292 | -0.2873 | No |
| 31 | Tgfbr1 | 10649 | -0.341 | -0.3077 | No |
| 32 | Bcar3 | 11044 | -0.393 | -0.3174 | Yes |
| 33 | Ppm1a | 11244 | -0.425 | -0.3095 | Yes |
| 34 | Ifngr2 | 11246 | -0.425 | -0.2855 | Yes |
| 35 | Ppp1ca | 11507 | -0.469 | -0.2800 | Yes |
| 36 | Rab31 | 11699 | -0.507 | -0.2667 | Yes |
| 37 | Trim33 | 11748 | -0.518 | -0.2413 | Yes |
| 38 | Fkbp1a | 11749 | -0.518 | -0.2119 | Yes |
| 39 | Apc | 11775 | -0.525 | -0.1842 | Yes |
| 40 | Slc20a1 | 11856 | -0.547 | -0.1597 | Yes |
| 41 | Cdk9 | 12058 | -0.619 | -0.1409 | Yes |
| 42 | Rhoa | 12123 | -0.658 | -0.1088 | Yes |
| 43 | Hdac1 | 12190 | -0.706 | -0.0741 | Yes |
| 44 | Furin | 12227 | -0.740 | -0.0351 | Yes |
| 45 | Smad3 | 12287 | -0.813 | 0.0062 | Yes |