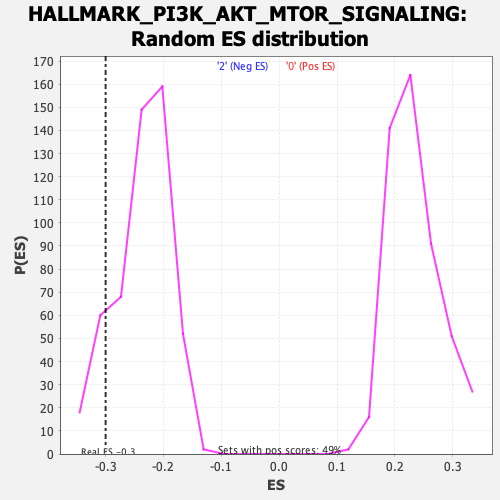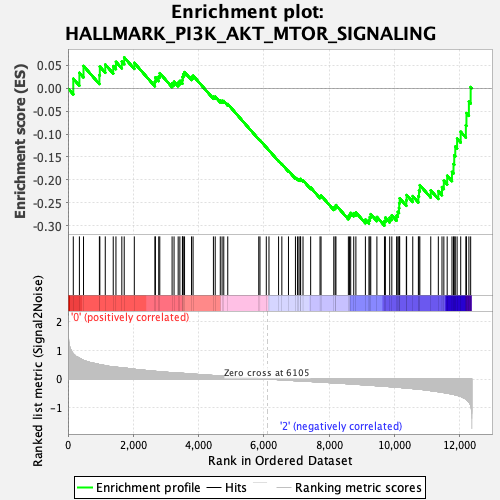
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.2999133 |
| Normalized Enrichment Score (NES) | -1.2788097 |
| Nominal p-value | 0.09448819 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.856 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Traf2 | 162 | 0.874 | 0.0210 | No |
| 2 | Stat2 | 349 | 0.718 | 0.0340 | No |
| 3 | Grb2 | 475 | 0.641 | 0.0489 | No |
| 4 | Arhgdia | 961 | 0.501 | 0.0289 | No |
| 5 | Ddit3 | 975 | 0.497 | 0.0473 | No |
| 6 | Il2rg | 1141 | 0.461 | 0.0520 | No |
| 7 | Ube2d3 | 1385 | 0.419 | 0.0485 | No |
| 8 | Hsp90b1 | 1468 | 0.410 | 0.0579 | No |
| 9 | Il4 | 1648 | 0.388 | 0.0585 | No |
| 10 | Mapk9 | 1721 | 0.380 | 0.0675 | No |
| 11 | Cltc | 2030 | 0.337 | 0.0556 | No |
| 12 | Ywhab | 2661 | 0.266 | 0.0147 | No |
| 13 | Raf1 | 2676 | 0.265 | 0.0239 | No |
| 14 | Mapkap1 | 2778 | 0.256 | 0.0257 | No |
| 15 | Arf1 | 2810 | 0.252 | 0.0331 | No |
| 16 | Ube2n | 3187 | 0.220 | 0.0111 | No |
| 17 | Akt1 | 3247 | 0.215 | 0.0147 | No |
| 18 | Pdk1 | 3373 | 0.205 | 0.0125 | No |
| 19 | Map2k3 | 3425 | 0.203 | 0.0163 | No |
| 20 | Cdkn1b | 3503 | 0.197 | 0.0177 | No |
| 21 | Cdk2 | 3508 | 0.196 | 0.0251 | No |
| 22 | Mknk2 | 3527 | 0.194 | 0.0312 | No |
| 23 | Pak4 | 3568 | 0.191 | 0.0354 | No |
| 24 | Mapk1 | 3782 | 0.174 | 0.0249 | No |
| 25 | Acaca | 3829 | 0.169 | 0.0278 | No |
| 26 | Them4 | 4452 | 0.120 | -0.0182 | No |
| 27 | Smad2 | 4507 | 0.115 | -0.0181 | No |
| 28 | Rps6ka3 | 4662 | 0.102 | -0.0267 | No |
| 29 | Arpc3 | 4717 | 0.097 | -0.0273 | No |
| 30 | Myd88 | 4769 | 0.094 | -0.0278 | No |
| 31 | Cab39l | 4891 | 0.085 | -0.0343 | No |
| 32 | Mapk8 | 5840 | 0.017 | -0.1109 | No |
| 33 | Actr2 | 5877 | 0.014 | -0.1133 | No |
| 34 | Atf1 | 6071 | 0.002 | -0.1289 | No |
| 35 | Prkaa2 | 6153 | -0.001 | -0.1355 | No |
| 36 | Vav3 | 6447 | -0.022 | -0.1585 | No |
| 37 | Plcg1 | 6547 | -0.029 | -0.1655 | No |
| 38 | Pten | 6750 | -0.042 | -0.1803 | No |
| 39 | Cab39 | 6963 | -0.057 | -0.1953 | No |
| 40 | Lck | 7025 | -0.061 | -0.1979 | No |
| 41 | Sla | 7070 | -0.063 | -0.1990 | No |
| 42 | Sqstm1 | 7109 | -0.066 | -0.1996 | No |
| 43 | Ralb | 7122 | -0.067 | -0.1979 | No |
| 44 | Actr3 | 7195 | -0.070 | -0.2011 | No |
| 45 | Rit1 | 7429 | -0.084 | -0.2167 | No |
| 46 | Tbk1 | 7716 | -0.106 | -0.2359 | No |
| 47 | Slc2a1 | 7747 | -0.108 | -0.2341 | No |
| 48 | E2f1 | 8137 | -0.133 | -0.2606 | No |
| 49 | Ppp2r1b | 8179 | -0.136 | -0.2586 | No |
| 50 | Ap2m1 | 8203 | -0.138 | -0.2551 | No |
| 51 | Calr | 8584 | -0.163 | -0.2797 | No |
| 52 | Map3k7 | 8620 | -0.166 | -0.2761 | No |
| 53 | Pin1 | 8655 | -0.168 | -0.2722 | No |
| 54 | Eif4e | 8748 | -0.176 | -0.2728 | No |
| 55 | Tiam1 | 8814 | -0.181 | -0.2711 | No |
| 56 | Itpr2 | 9105 | -0.203 | -0.2867 | No |
| 57 | Plcb1 | 9214 | -0.210 | -0.2873 | No |
| 58 | Map2k6 | 9242 | -0.212 | -0.2812 | No |
| 59 | Rac1 | 9269 | -0.215 | -0.2749 | No |
| 60 | Ecsit | 9457 | -0.231 | -0.2811 | No |
| 61 | Dapp1 | 9689 | -0.252 | -0.2901 | Yes |
| 62 | Cxcr4 | 9717 | -0.253 | -0.2823 | Yes |
| 63 | Akt1s1 | 9849 | -0.263 | -0.2827 | Yes |
| 64 | Ptpn11 | 9914 | -0.270 | -0.2774 | Yes |
| 65 | Grk2 | 10060 | -0.284 | -0.2781 | Yes |
| 66 | Gna14 | 10098 | -0.286 | -0.2699 | Yes |
| 67 | Pfn1 | 10133 | -0.288 | -0.2614 | Yes |
| 68 | Nod1 | 10141 | -0.290 | -0.2506 | Yes |
| 69 | Tsc2 | 10155 | -0.291 | -0.2403 | Yes |
| 70 | Mknk1 | 10358 | -0.310 | -0.2446 | Yes |
| 71 | Cdk1 | 10367 | -0.310 | -0.2331 | Yes |
| 72 | Prkcb | 10555 | -0.331 | -0.2354 | Yes |
| 73 | Prkar2a | 10727 | -0.349 | -0.2357 | Yes |
| 74 | Pikfyve | 10746 | -0.351 | -0.2234 | Yes |
| 75 | Irak4 | 10772 | -0.354 | -0.2116 | Yes |
| 76 | Dusp3 | 11106 | -0.402 | -0.2230 | Yes |
| 77 | Pla2g12a | 11338 | -0.438 | -0.2246 | Yes |
| 78 | Nfkbib | 11450 | -0.460 | -0.2156 | Yes |
| 79 | Ppp1ca | 11507 | -0.469 | -0.2018 | Yes |
| 80 | Tnfrsf1a | 11609 | -0.489 | -0.1909 | Yes |
| 81 | Csnk2b | 11756 | -0.520 | -0.1825 | Yes |
| 82 | Rps6ka1 | 11803 | -0.532 | -0.1654 | Yes |
| 83 | Nck1 | 11825 | -0.537 | -0.1461 | Yes |
| 84 | Cfl1 | 11857 | -0.547 | -0.1272 | Yes |
| 85 | Gsk3b | 11913 | -0.562 | -0.1097 | Yes |
| 86 | Trib3 | 12023 | -0.604 | -0.0949 | Yes |
| 87 | Rptor | 12181 | -0.691 | -0.0807 | Yes |
| 88 | Prkag1 | 12199 | -0.714 | -0.0541 | Yes |
| 89 | Ripk1 | 12274 | -0.794 | -0.0291 | Yes |
| 90 | Cdk4 | 12328 | -0.926 | 0.0029 | Yes |