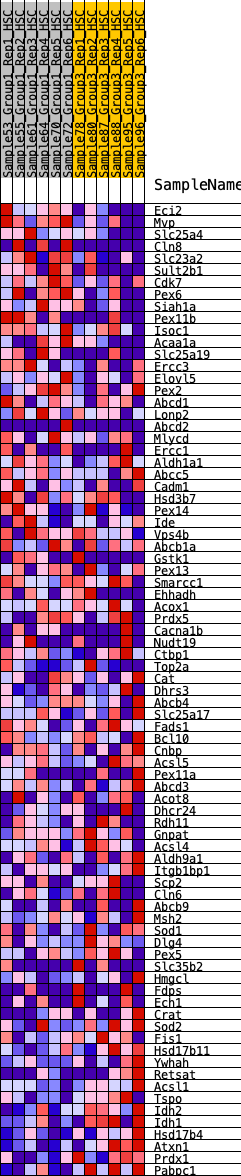
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
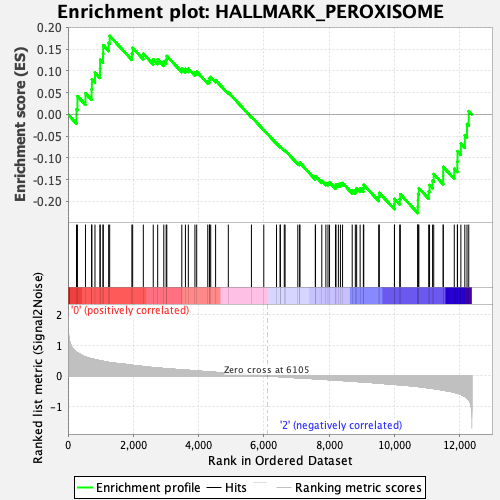
| GeneSet | HALLMARK_PEROXISOME |
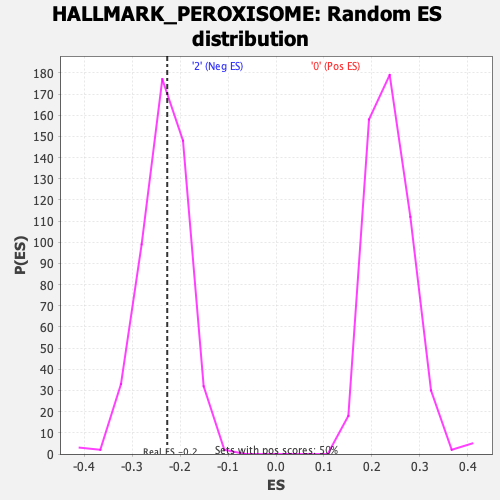
| Enrichment Score (ES) | -0.22687754 |
| Normalized Enrichment Score (NES) | -0.9631313 |
| Nominal p-value | 0.5645161 |
| FDR q-value | 0.73846185 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eci2 | 260 | 0.773 | 0.0120 | No |
| 2 | Mvp | 289 | 0.757 | 0.0421 | No |
| 3 | Slc25a4 | 535 | 0.619 | 0.0487 | No |
| 4 | Cln8 | 722 | 0.560 | 0.0576 | No |
| 5 | Slc23a2 | 731 | 0.559 | 0.0808 | No |
| 6 | Sult2b1 | 826 | 0.536 | 0.0962 | No |
| 7 | Cdk7 | 980 | 0.496 | 0.1050 | No |
| 8 | Pex6 | 985 | 0.495 | 0.1258 | No |
| 9 | Siah1a | 1071 | 0.476 | 0.1393 | No |
| 10 | Pex11b | 1078 | 0.475 | 0.1592 | No |
| 11 | Isoc1 | 1245 | 0.440 | 0.1645 | No |
| 12 | Acaa1a | 1276 | 0.433 | 0.1806 | No |
| 13 | Slc25a19 | 1956 | 0.348 | 0.1402 | No |
| 14 | Ercc3 | 1980 | 0.344 | 0.1531 | No |
| 15 | Elovl5 | 2306 | 0.305 | 0.1397 | No |
| 16 | Pex2 | 2610 | 0.270 | 0.1266 | No |
| 17 | Abcd1 | 2746 | 0.259 | 0.1267 | No |
| 18 | Lonp2 | 2933 | 0.240 | 0.1218 | No |
| 19 | Abcd2 | 3005 | 0.233 | 0.1260 | No |
| 20 | Mlycd | 3025 | 0.231 | 0.1344 | No |
| 21 | Ercc1 | 3484 | 0.198 | 0.1056 | No |
| 22 | Aldh1a1 | 3595 | 0.189 | 0.1047 | No |
| 23 | Abcc5 | 3684 | 0.182 | 0.1053 | No |
| 24 | Cadm1 | 3886 | 0.164 | 0.0960 | No |
| 25 | Hsd3b7 | 3940 | 0.160 | 0.0985 | No |
| 26 | Pex14 | 4279 | 0.133 | 0.0767 | No |
| 27 | Ide | 4330 | 0.129 | 0.0781 | No |
| 28 | Vps4b | 4334 | 0.128 | 0.0834 | No |
| 29 | Abcb1a | 4369 | 0.127 | 0.0860 | No |
| 30 | Gstk1 | 4519 | 0.114 | 0.0788 | No |
| 31 | Pex13 | 4908 | 0.084 | 0.0508 | No |
| 32 | Smarcc1 | 5616 | 0.033 | -0.0053 | No |
| 33 | Ehhadh | 5994 | 0.007 | -0.0357 | No |
| 34 | Acox1 | 6384 | -0.017 | -0.0667 | No |
| 35 | Prdx5 | 6495 | -0.025 | -0.0746 | No |
| 36 | Cacna1b | 6500 | -0.025 | -0.0738 | No |
| 37 | Nudt19 | 6622 | -0.033 | -0.0823 | No |
| 38 | Ctbp1 | 6646 | -0.035 | -0.0826 | No |
| 39 | Top2a | 7036 | -0.061 | -0.1117 | No |
| 40 | Cat | 7096 | -0.065 | -0.1137 | No |
| 41 | Dhrs3 | 7097 | -0.065 | -0.1109 | No |
| 42 | Abcb4 | 7571 | -0.095 | -0.1454 | No |
| 43 | Slc25a17 | 7578 | -0.096 | -0.1417 | No |
| 44 | Fads1 | 7770 | -0.109 | -0.1526 | No |
| 45 | Bcl10 | 7895 | -0.117 | -0.1577 | No |
| 46 | Cnbp | 7960 | -0.121 | -0.1577 | No |
| 47 | Acsl5 | 8007 | -0.123 | -0.1562 | No |
| 48 | Pex11a | 8189 | -0.137 | -0.1651 | No |
| 49 | Abcd3 | 8205 | -0.138 | -0.1604 | No |
| 50 | Acot8 | 8278 | -0.144 | -0.1601 | No |
| 51 | Dhcr24 | 8341 | -0.148 | -0.1588 | No |
| 52 | Rdh11 | 8406 | -0.152 | -0.1575 | No |
| 53 | Gnpat | 8701 | -0.172 | -0.1741 | No |
| 54 | Acsl4 | 8797 | -0.180 | -0.1741 | No |
| 55 | Aldh9a1 | 8837 | -0.183 | -0.1695 | No |
| 56 | Itgb1bp1 | 8944 | -0.192 | -0.1699 | No |
| 57 | Scp2 | 9043 | -0.198 | -0.1694 | No |
| 58 | Cln6 | 9055 | -0.199 | -0.1617 | No |
| 59 | Abcb9 | 9513 | -0.236 | -0.1888 | No |
| 60 | Msh2 | 9531 | -0.237 | -0.1801 | No |
| 61 | Sod1 | 9993 | -0.277 | -0.2057 | No |
| 62 | Dlg4 | 9996 | -0.277 | -0.1940 | No |
| 63 | Pex5 | 10156 | -0.291 | -0.1945 | No |
| 64 | Slc35b2 | 10174 | -0.293 | -0.1833 | No |
| 65 | Hmgcl | 10710 | -0.348 | -0.2120 | Yes |
| 66 | Fdps | 10714 | -0.348 | -0.1973 | Yes |
| 67 | Ech1 | 10718 | -0.348 | -0.1826 | Yes |
| 68 | Crat | 10742 | -0.351 | -0.1694 | Yes |
| 69 | Sod2 | 11043 | -0.392 | -0.1771 | Yes |
| 70 | Fis1 | 11069 | -0.397 | -0.1621 | Yes |
| 71 | Hsd17b11 | 11162 | -0.411 | -0.1520 | Yes |
| 72 | Ywhah | 11197 | -0.416 | -0.1369 | Yes |
| 73 | Retsat | 11487 | -0.466 | -0.1405 | Yes |
| 74 | Acsl1 | 11488 | -0.466 | -0.1205 | Yes |
| 75 | Tspo | 11826 | -0.538 | -0.1249 | Yes |
| 76 | Idh2 | 11920 | -0.565 | -0.1083 | Yes |
| 77 | Idh1 | 11926 | -0.566 | -0.0845 | Yes |
| 78 | Hsd17b4 | 12028 | -0.604 | -0.0668 | Yes |
| 79 | Atxn1 | 12149 | -0.669 | -0.0479 | Yes |
| 80 | Prdx1 | 12216 | -0.731 | -0.0219 | Yes |
| 81 | Pabpc1 | 12269 | -0.790 | 0.0077 | Yes |