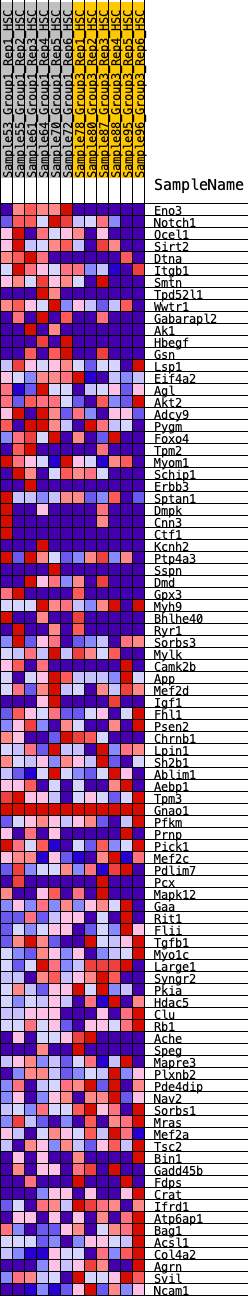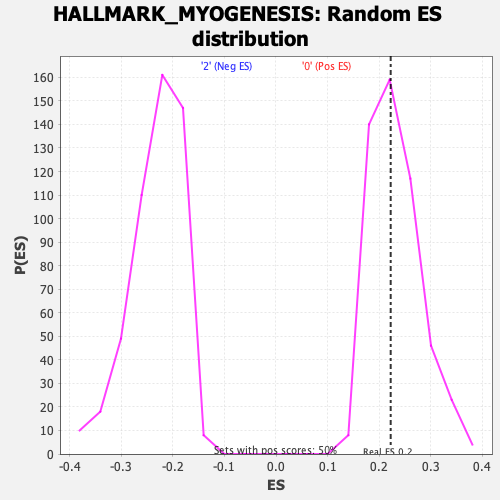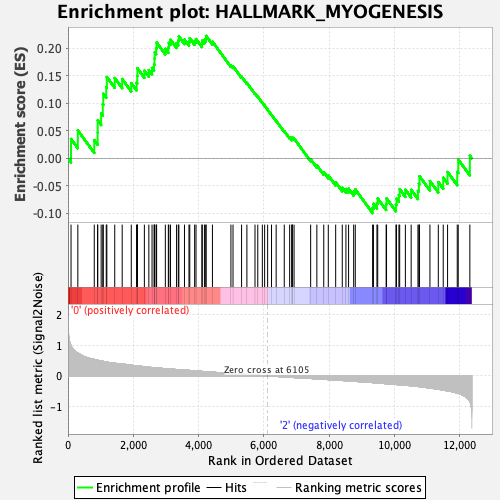
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.22234565 |
| Normalized Enrichment Score (NES) | 0.9599632 |
| Nominal p-value | 0.52716297 |
| FDR q-value | 0.88942957 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eno3 | 93 | 0.993 | 0.0352 | Yes |
| 2 | Notch1 | 301 | 0.752 | 0.0507 | Yes |
| 3 | Ocel1 | 804 | 0.540 | 0.0331 | Yes |
| 4 | Sirt2 | 907 | 0.513 | 0.0469 | Yes |
| 5 | Dtna | 909 | 0.513 | 0.0689 | Yes |
| 6 | Itgb1 | 1015 | 0.490 | 0.0814 | Yes |
| 7 | Smtn | 1065 | 0.477 | 0.0980 | Yes |
| 8 | Tpd52l1 | 1080 | 0.475 | 0.1173 | Yes |
| 9 | Wwtr1 | 1171 | 0.456 | 0.1296 | Yes |
| 10 | Gabarapl2 | 1189 | 0.452 | 0.1477 | Yes |
| 11 | Ak1 | 1431 | 0.413 | 0.1459 | Yes |
| 12 | Hbegf | 1659 | 0.385 | 0.1440 | Yes |
| 13 | Gsn | 1937 | 0.350 | 0.1365 | Yes |
| 14 | Lsp1 | 2099 | 0.328 | 0.1375 | Yes |
| 15 | Eif4a2 | 2121 | 0.325 | 0.1498 | Yes |
| 16 | Agl | 2126 | 0.324 | 0.1634 | Yes |
| 17 | Akt2 | 2338 | 0.301 | 0.1592 | Yes |
| 18 | Adcy9 | 2475 | 0.283 | 0.1604 | Yes |
| 19 | Pygm | 2573 | 0.275 | 0.1643 | Yes |
| 20 | Foxo4 | 2635 | 0.268 | 0.1709 | Yes |
| 21 | Tpm2 | 2649 | 0.267 | 0.1813 | Yes |
| 22 | Myom1 | 2654 | 0.267 | 0.1925 | Yes |
| 23 | Schip1 | 2700 | 0.262 | 0.2001 | Yes |
| 24 | Erbb3 | 2713 | 0.261 | 0.2104 | Yes |
| 25 | Sptan1 | 2980 | 0.234 | 0.1988 | Yes |
| 26 | Dmpk | 3072 | 0.227 | 0.2012 | Yes |
| 27 | Cnn3 | 3087 | 0.226 | 0.2098 | Yes |
| 28 | Ctf1 | 3137 | 0.224 | 0.2154 | Yes |
| 29 | Kcnh2 | 3324 | 0.209 | 0.2093 | Yes |
| 30 | Ptp4a3 | 3377 | 0.205 | 0.2139 | Yes |
| 31 | Sspn | 3394 | 0.204 | 0.2213 | Yes |
| 32 | Dmd | 3566 | 0.191 | 0.2156 | Yes |
| 33 | Gpx3 | 3704 | 0.180 | 0.2122 | Yes |
| 34 | Myh9 | 3728 | 0.178 | 0.2180 | Yes |
| 35 | Bhlhe40 | 3877 | 0.165 | 0.2131 | Yes |
| 36 | Ryr1 | 3919 | 0.162 | 0.2167 | Yes |
| 37 | Sorbs3 | 4100 | 0.146 | 0.2083 | Yes |
| 38 | Mylk | 4110 | 0.146 | 0.2139 | Yes |
| 39 | Camk2b | 4177 | 0.140 | 0.2145 | Yes |
| 40 | App | 4218 | 0.137 | 0.2172 | Yes |
| 41 | Mef2d | 4228 | 0.137 | 0.2223 | Yes |
| 42 | Igf1 | 4422 | 0.123 | 0.2119 | No |
| 43 | Fhl1 | 4988 | 0.078 | 0.1692 | No |
| 44 | Psen2 | 5052 | 0.073 | 0.1672 | No |
| 45 | Chrnb1 | 5314 | 0.054 | 0.1483 | No |
| 46 | Lpin1 | 5477 | 0.042 | 0.1369 | No |
| 47 | Sh2b1 | 5725 | 0.026 | 0.1179 | No |
| 48 | Ablim1 | 5810 | 0.020 | 0.1120 | No |
| 49 | Aebp1 | 5950 | 0.010 | 0.1010 | No |
| 50 | Tpm3 | 6020 | 0.005 | 0.0956 | No |
| 51 | Gnao1 | 6112 | 0.000 | 0.0882 | No |
| 52 | Pfkm | 6231 | -0.006 | 0.0789 | No |
| 53 | Prnp | 6373 | -0.016 | 0.0681 | No |
| 54 | Pick1 | 6621 | -0.033 | 0.0494 | No |
| 55 | Mef2c | 6786 | -0.044 | 0.0379 | No |
| 56 | Pdlim7 | 6851 | -0.049 | 0.0348 | No |
| 57 | Pcx | 6862 | -0.050 | 0.0362 | No |
| 58 | Mapk12 | 6866 | -0.050 | 0.0381 | No |
| 59 | Gaa | 6921 | -0.054 | 0.0360 | No |
| 60 | Rit1 | 7429 | -0.084 | -0.0016 | No |
| 61 | Flii | 7619 | -0.099 | -0.0128 | No |
| 62 | Tgfb1 | 7830 | -0.113 | -0.0250 | No |
| 63 | Myo1c | 7969 | -0.122 | -0.0310 | No |
| 64 | Large1 | 8195 | -0.137 | -0.0434 | No |
| 65 | Syngr2 | 8397 | -0.151 | -0.0533 | No |
| 66 | Pkia | 8509 | -0.158 | -0.0555 | No |
| 67 | Hdac5 | 8590 | -0.163 | -0.0550 | No |
| 68 | Clu | 8746 | -0.176 | -0.0601 | No |
| 69 | Rb1 | 8795 | -0.179 | -0.0563 | No |
| 70 | Ache | 9326 | -0.219 | -0.0900 | No |
| 71 | Speg | 9351 | -0.222 | -0.0824 | No |
| 72 | Mapre3 | 9459 | -0.231 | -0.0811 | No |
| 73 | Plxnb2 | 9482 | -0.233 | -0.0729 | No |
| 74 | Pde4dip | 9740 | -0.255 | -0.0828 | No |
| 75 | Nav2 | 9752 | -0.256 | -0.0727 | No |
| 76 | Sorbs1 | 10040 | -0.281 | -0.0840 | No |
| 77 | Mras | 10058 | -0.283 | -0.0731 | No |
| 78 | Mef2a | 10131 | -0.288 | -0.0666 | No |
| 79 | Tsc2 | 10155 | -0.291 | -0.0559 | No |
| 80 | Bin1 | 10329 | -0.307 | -0.0568 | No |
| 81 | Gadd45b | 10506 | -0.326 | -0.0571 | No |
| 82 | Fdps | 10714 | -0.348 | -0.0590 | No |
| 83 | Crat | 10742 | -0.351 | -0.0460 | No |
| 84 | Ifrd1 | 10762 | -0.353 | -0.0324 | No |
| 85 | Atp6ap1 | 11081 | -0.398 | -0.0411 | No |
| 86 | Bag1 | 11337 | -0.438 | -0.0430 | No |
| 87 | Acsl1 | 11488 | -0.466 | -0.0352 | No |
| 88 | Col4a2 | 11621 | -0.492 | -0.0247 | No |
| 89 | Agrn | 11917 | -0.564 | -0.0245 | No |
| 90 | Svil | 11947 | -0.575 | -0.0020 | No |
| 91 | Ncam1 | 12303 | -0.832 | 0.0049 | No |