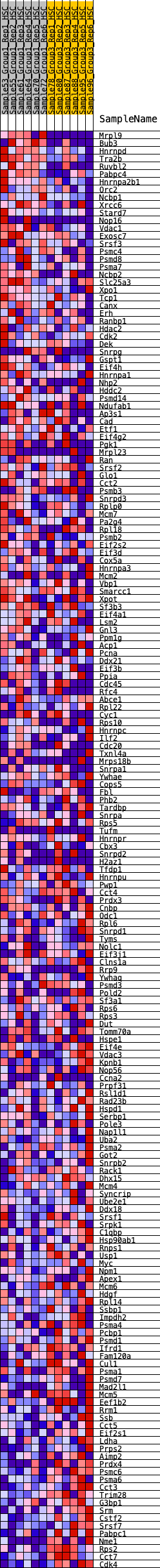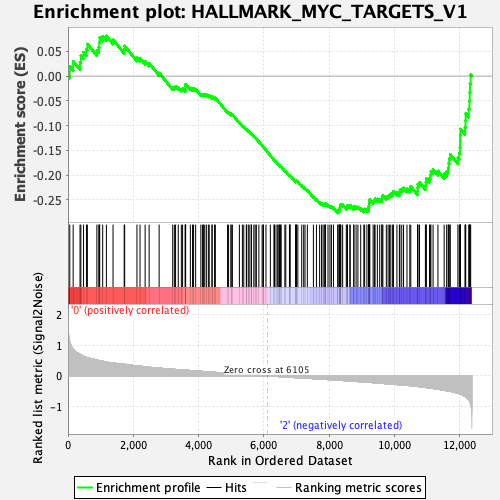
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYC_TARGETS_V1 |
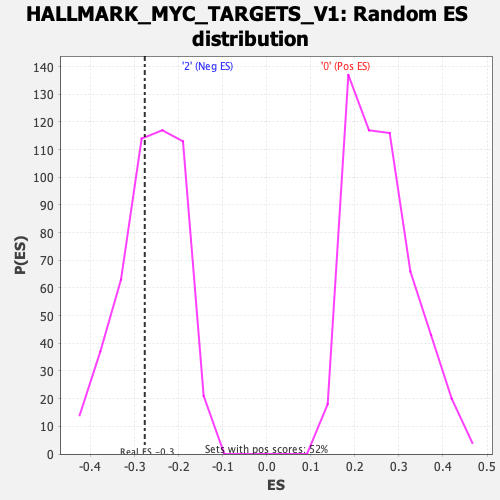
| Enrichment Score (ES) | -0.27681696 |
| Normalized Enrichment Score (NES) | -1.0571096 |
| Nominal p-value | 0.39039665 |
| FDR q-value | 0.72539914 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mrpl9 | 54 | 1.108 | 0.0195 | No |
| 2 | Bub3 | 158 | 0.877 | 0.0299 | No |
| 3 | Hnrnpd | 368 | 0.708 | 0.0280 | No |
| 4 | Tra2b | 394 | 0.696 | 0.0410 | No |
| 5 | Ruvbl2 | 476 | 0.641 | 0.0481 | No |
| 6 | Pabpc4 | 565 | 0.607 | 0.0540 | No |
| 7 | Hnrnpa2b1 | 593 | 0.598 | 0.0647 | No |
| 8 | Orc2 | 884 | 0.519 | 0.0521 | No |
| 9 | Ncbp1 | 944 | 0.504 | 0.0581 | No |
| 10 | Xrcc6 | 958 | 0.501 | 0.0678 | No |
| 11 | Stard7 | 971 | 0.498 | 0.0776 | No |
| 12 | Nop16 | 1062 | 0.477 | 0.0805 | No |
| 13 | Vdac1 | 1175 | 0.456 | 0.0811 | No |
| 14 | Exosc7 | 1379 | 0.420 | 0.0735 | No |
| 15 | Srsf3 | 1720 | 0.380 | 0.0538 | No |
| 16 | Psmc4 | 1735 | 0.378 | 0.0608 | No |
| 17 | Psmd8 | 2111 | 0.326 | 0.0370 | No |
| 18 | Psma7 | 2205 | 0.317 | 0.0362 | No |
| 19 | Ncbp2 | 2361 | 0.298 | 0.0299 | No |
| 20 | Slc25a3 | 2484 | 0.282 | 0.0260 | No |
| 21 | Xpo1 | 2790 | 0.255 | 0.0064 | No |
| 22 | Tcp1 | 3206 | 0.219 | -0.0229 | No |
| 23 | Canx | 3256 | 0.214 | -0.0223 | No |
| 24 | Erh | 3293 | 0.210 | -0.0208 | No |
| 25 | Ranbp1 | 3376 | 0.205 | -0.0231 | No |
| 26 | Hdac2 | 3470 | 0.199 | -0.0264 | No |
| 27 | Cdk2 | 3508 | 0.196 | -0.0252 | No |
| 28 | Dek | 3586 | 0.190 | -0.0275 | No |
| 29 | Snrpg | 3596 | 0.189 | -0.0241 | No |
| 30 | Gspt1 | 3597 | 0.189 | -0.0201 | No |
| 31 | Eif4h | 3602 | 0.188 | -0.0163 | No |
| 32 | Hnrnpa1 | 3746 | 0.176 | -0.0243 | No |
| 33 | Nhp2 | 3813 | 0.171 | -0.0260 | No |
| 34 | Hddc2 | 3844 | 0.168 | -0.0249 | No |
| 35 | Psmd14 | 3908 | 0.162 | -0.0265 | No |
| 36 | Ndufab1 | 4068 | 0.149 | -0.0364 | No |
| 37 | Ap3s1 | 4120 | 0.145 | -0.0374 | No |
| 38 | Cad | 4146 | 0.143 | -0.0364 | No |
| 39 | Etf1 | 4186 | 0.140 | -0.0366 | No |
| 40 | Eif4g2 | 4236 | 0.136 | -0.0377 | No |
| 41 | Pgk1 | 4297 | 0.131 | -0.0398 | No |
| 42 | Mrpl23 | 4323 | 0.129 | -0.0390 | No |
| 43 | Ran | 4410 | 0.123 | -0.0434 | No |
| 44 | Srsf2 | 4411 | 0.123 | -0.0408 | No |
| 45 | Glo1 | 4487 | 0.117 | -0.0444 | No |
| 46 | Cct2 | 4511 | 0.115 | -0.0438 | No |
| 47 | Psmb3 | 4885 | 0.085 | -0.0726 | No |
| 48 | Snrpd3 | 4916 | 0.083 | -0.0733 | No |
| 49 | Rplp0 | 4991 | 0.078 | -0.0777 | No |
| 50 | Mcm7 | 4993 | 0.078 | -0.0761 | No |
| 51 | Pa2g4 | 5033 | 0.074 | -0.0777 | No |
| 52 | Rpl18 | 5247 | 0.059 | -0.0939 | No |
| 53 | Psmb2 | 5339 | 0.052 | -0.1003 | No |
| 54 | Eif2s2 | 5381 | 0.049 | -0.1026 | No |
| 55 | Eif3d | 5460 | 0.044 | -0.1080 | No |
| 56 | Cox5a | 5464 | 0.044 | -0.1073 | No |
| 57 | Hnrnpa3 | 5518 | 0.040 | -0.1108 | No |
| 58 | Mcm2 | 5546 | 0.038 | -0.1122 | No |
| 59 | Vbp1 | 5606 | 0.034 | -0.1163 | No |
| 60 | Smarcc1 | 5616 | 0.033 | -0.1164 | No |
| 61 | Xpot | 5689 | 0.028 | -0.1217 | No |
| 62 | Sf3b3 | 5730 | 0.026 | -0.1244 | No |
| 63 | Eif4a1 | 5767 | 0.023 | -0.1269 | No |
| 64 | Lsm2 | 5843 | 0.017 | -0.1327 | No |
| 65 | Gnl3 | 5943 | 0.010 | -0.1406 | No |
| 66 | Ppm1g | 5972 | 0.008 | -0.1427 | No |
| 67 | Acp1 | 5985 | 0.008 | -0.1435 | No |
| 68 | Pcna | 6061 | 0.002 | -0.1496 | No |
| 69 | Ddx21 | 6195 | -0.003 | -0.1605 | No |
| 70 | Eif3b | 6305 | -0.011 | -0.1692 | No |
| 71 | Ppia | 6323 | -0.012 | -0.1703 | No |
| 72 | Cdc45 | 6369 | -0.016 | -0.1737 | No |
| 73 | Rfc4 | 6423 | -0.020 | -0.1776 | No |
| 74 | Abce1 | 6433 | -0.021 | -0.1779 | No |
| 75 | Rpl22 | 6474 | -0.024 | -0.1807 | No |
| 76 | Cyc1 | 6488 | -0.025 | -0.1812 | No |
| 77 | Rps10 | 6503 | -0.025 | -0.1818 | No |
| 78 | Hnrnpc | 6527 | -0.027 | -0.1831 | No |
| 79 | Ilf2 | 6639 | -0.035 | -0.1915 | No |
| 80 | Cdc20 | 6668 | -0.037 | -0.1930 | No |
| 81 | Txnl4a | 6780 | -0.044 | -0.2011 | No |
| 82 | Mrps18b | 6782 | -0.044 | -0.2003 | No |
| 83 | Snrpa1 | 6803 | -0.045 | -0.2009 | No |
| 84 | Ywhae | 6968 | -0.057 | -0.2132 | No |
| 85 | Cops5 | 6976 | -0.058 | -0.2125 | No |
| 86 | Fbl | 6989 | -0.059 | -0.2122 | No |
| 87 | Phb2 | 6993 | -0.059 | -0.2112 | No |
| 88 | Tardbp | 7037 | -0.061 | -0.2134 | No |
| 89 | Snrpa | 7156 | -0.069 | -0.2216 | No |
| 90 | Rps5 | 7216 | -0.071 | -0.2249 | No |
| 91 | Tufm | 7256 | -0.075 | -0.2265 | No |
| 92 | Hnrnpr | 7328 | -0.079 | -0.2306 | No |
| 93 | Cbx3 | 7513 | -0.091 | -0.2438 | No |
| 94 | Snrpd2 | 7606 | -0.098 | -0.2492 | No |
| 95 | H2az1 | 7704 | -0.104 | -0.2549 | No |
| 96 | Tfdp1 | 7763 | -0.108 | -0.2574 | No |
| 97 | Hnrnpu | 7809 | -0.112 | -0.2586 | No |
| 98 | Pwp1 | 7848 | -0.114 | -0.2593 | No |
| 99 | Cct4 | 7880 | -0.116 | -0.2593 | No |
| 100 | Prdx3 | 7882 | -0.116 | -0.2569 | No |
| 101 | Cnbp | 7960 | -0.121 | -0.2606 | No |
| 102 | Odc1 | 8020 | -0.124 | -0.2628 | No |
| 103 | Rpl6 | 8064 | -0.128 | -0.2636 | No |
| 104 | Snrpd1 | 8127 | -0.132 | -0.2658 | No |
| 105 | Tyms | 8262 | -0.143 | -0.2737 | Yes |
| 106 | Nolc1 | 8263 | -0.143 | -0.2707 | Yes |
| 107 | Eif3j1 | 8306 | -0.145 | -0.2710 | Yes |
| 108 | Clns1a | 8308 | -0.145 | -0.2679 | Yes |
| 109 | Rrp9 | 8326 | -0.147 | -0.2662 | Yes |
| 110 | Ywhaq | 8330 | -0.147 | -0.2632 | Yes |
| 111 | Psmd3 | 8333 | -0.147 | -0.2602 | Yes |
| 112 | Pold2 | 8355 | -0.149 | -0.2587 | Yes |
| 113 | Sf3a1 | 8409 | -0.152 | -0.2598 | Yes |
| 114 | Rps6 | 8520 | -0.159 | -0.2654 | Yes |
| 115 | Rps3 | 8544 | -0.160 | -0.2639 | Yes |
| 116 | Dut | 8555 | -0.161 | -0.2612 | Yes |
| 117 | Tomm70a | 8629 | -0.166 | -0.2636 | Yes |
| 118 | Hspe1 | 8639 | -0.167 | -0.2608 | Yes |
| 119 | Eif4e | 8748 | -0.176 | -0.2658 | Yes |
| 120 | Vdac3 | 8755 | -0.177 | -0.2625 | Yes |
| 121 | Kpnb1 | 8820 | -0.181 | -0.2639 | Yes |
| 122 | Nop56 | 8876 | -0.186 | -0.2644 | Yes |
| 123 | Ccna2 | 8961 | -0.193 | -0.2671 | Yes |
| 124 | Prpf31 | 9057 | -0.199 | -0.2706 | Yes |
| 125 | Rsl1d1 | 9082 | -0.201 | -0.2682 | Yes |
| 126 | Rad23b | 9151 | -0.207 | -0.2694 | Yes |
| 127 | Hspd1 | 9201 | -0.210 | -0.2689 | Yes |
| 128 | Serbp1 | 9202 | -0.210 | -0.2644 | Yes |
| 129 | Pole3 | 9216 | -0.210 | -0.2609 | Yes |
| 130 | Nap1l1 | 9224 | -0.211 | -0.2569 | Yes |
| 131 | Uba2 | 9229 | -0.211 | -0.2527 | Yes |
| 132 | Psma2 | 9240 | -0.212 | -0.2489 | Yes |
| 133 | Got2 | 9342 | -0.221 | -0.2525 | Yes |
| 134 | Snrpb2 | 9380 | -0.224 | -0.2507 | Yes |
| 135 | Rack1 | 9401 | -0.227 | -0.2474 | Yes |
| 136 | Dhx15 | 9471 | -0.232 | -0.2481 | Yes |
| 137 | Mcm4 | 9543 | -0.238 | -0.2488 | Yes |
| 138 | Syncrip | 9606 | -0.244 | -0.2486 | Yes |
| 139 | Ube2e1 | 9615 | -0.245 | -0.2440 | Yes |
| 140 | Ddx18 | 9640 | -0.247 | -0.2406 | Yes |
| 141 | Srsf1 | 9746 | -0.256 | -0.2437 | Yes |
| 142 | Srpk1 | 9800 | -0.260 | -0.2425 | Yes |
| 143 | C1qbp | 9841 | -0.263 | -0.2401 | Yes |
| 144 | Hsp90ab1 | 9883 | -0.267 | -0.2377 | Yes |
| 145 | Rnps1 | 9939 | -0.273 | -0.2363 | Yes |
| 146 | Usp1 | 9964 | -0.274 | -0.2324 | Yes |
| 147 | Myc | 10070 | -0.284 | -0.2349 | Yes |
| 148 | Npm1 | 10145 | -0.290 | -0.2347 | Yes |
| 149 | Apex1 | 10162 | -0.292 | -0.2297 | Yes |
| 150 | Mcm6 | 10218 | -0.297 | -0.2278 | Yes |
| 151 | Hdgf | 10273 | -0.303 | -0.2257 | Yes |
| 152 | Rpl14 | 10378 | -0.311 | -0.2276 | Yes |
| 153 | Ssbp1 | 10464 | -0.320 | -0.2276 | Yes |
| 154 | Impdh2 | 10495 | -0.324 | -0.2231 | Yes |
| 155 | Psma4 | 10703 | -0.347 | -0.2326 | Yes |
| 156 | Pcbp1 | 10707 | -0.348 | -0.2254 | Yes |
| 157 | Psmd1 | 10712 | -0.348 | -0.2182 | Yes |
| 158 | Ifrd1 | 10762 | -0.353 | -0.2146 | Yes |
| 159 | Fam120a | 10942 | -0.377 | -0.2212 | Yes |
| 160 | Cul1 | 10958 | -0.380 | -0.2142 | Yes |
| 161 | Psma1 | 10970 | -0.382 | -0.2069 | Yes |
| 162 | Psmd7 | 11068 | -0.397 | -0.2063 | Yes |
| 163 | Mad2l1 | 11092 | -0.401 | -0.1995 | Yes |
| 164 | Mcm5 | 11110 | -0.403 | -0.1922 | Yes |
| 165 | Eef1b2 | 11177 | -0.414 | -0.1887 | Yes |
| 166 | Rrm1 | 11326 | -0.436 | -0.1915 | Yes |
| 167 | Ssb | 11516 | -0.471 | -0.1968 | Yes |
| 168 | Cct5 | 11593 | -0.486 | -0.1926 | Yes |
| 169 | Eif2s1 | 11640 | -0.496 | -0.1857 | Yes |
| 170 | Ldha | 11655 | -0.498 | -0.1761 | Yes |
| 171 | Prps2 | 11672 | -0.502 | -0.1666 | Yes |
| 172 | Aimp2 | 11705 | -0.508 | -0.1583 | Yes |
| 173 | Prdx4 | 11937 | -0.571 | -0.1649 | Yes |
| 174 | Psmc6 | 11980 | -0.586 | -0.1557 | Yes |
| 175 | Psma6 | 11999 | -0.593 | -0.1444 | Yes |
| 176 | Cct3 | 12005 | -0.597 | -0.1319 | Yes |
| 177 | Trim28 | 12006 | -0.597 | -0.1191 | Yes |
| 178 | G3bp1 | 12016 | -0.600 | -0.1069 | Yes |
| 179 | Srm | 12154 | -0.672 | -0.1036 | Yes |
| 180 | Cstf2 | 12171 | -0.685 | -0.0902 | Yes |
| 181 | Srsf7 | 12176 | -0.688 | -0.0757 | Yes |
| 182 | Pabpc1 | 12269 | -0.790 | -0.0662 | Yes |
| 183 | Nme1 | 12290 | -0.816 | -0.0502 | Yes |
| 184 | Rps2 | 12298 | -0.826 | -0.0330 | Yes |
| 185 | Cct7 | 12307 | -0.844 | -0.0154 | Yes |
| 186 | Cdk4 | 12328 | -0.926 | 0.0029 | Yes |