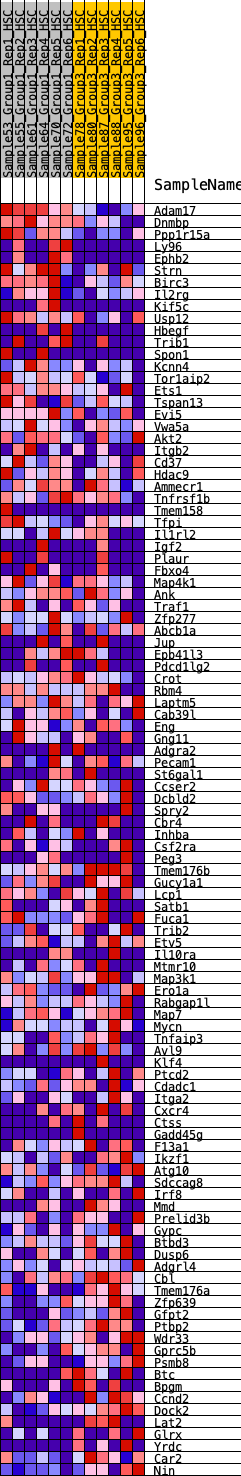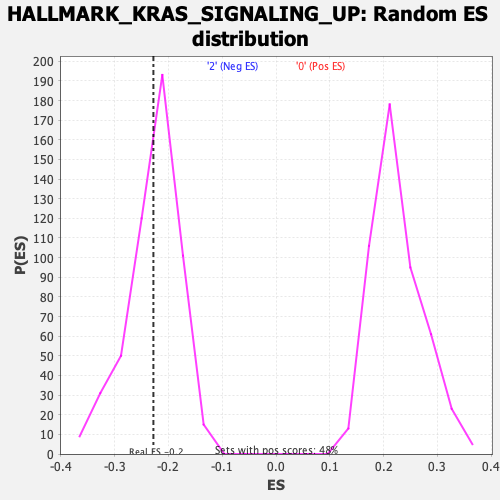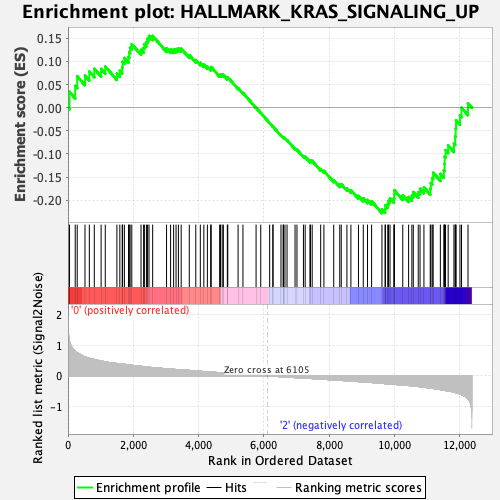
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.22773431 |
| Normalized Enrichment Score (NES) | -1.0035208 |
| Nominal p-value | 0.41811174 |
| FDR q-value | 0.661465 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adam17 | 47 | 1.133 | 0.0344 | No |
| 2 | Dnmbp | 221 | 0.798 | 0.0472 | No |
| 3 | Ppp1r15a | 282 | 0.760 | 0.0679 | No |
| 4 | Ly96 | 520 | 0.623 | 0.0696 | No |
| 5 | Ephb2 | 653 | 0.578 | 0.0783 | No |
| 6 | Strn | 805 | 0.540 | 0.0842 | No |
| 7 | Birc3 | 1014 | 0.490 | 0.0837 | No |
| 8 | Il2rg | 1141 | 0.461 | 0.0890 | No |
| 9 | Kif5c | 1497 | 0.406 | 0.0737 | No |
| 10 | Usp12 | 1587 | 0.396 | 0.0798 | No |
| 11 | Hbegf | 1659 | 0.385 | 0.0870 | No |
| 12 | Trib1 | 1671 | 0.385 | 0.0991 | No |
| 13 | Spon1 | 1726 | 0.379 | 0.1075 | No |
| 14 | Kcnn4 | 1853 | 0.363 | 0.1094 | No |
| 15 | Tor1aip2 | 1876 | 0.360 | 0.1198 | No |
| 16 | Ets1 | 1902 | 0.355 | 0.1297 | No |
| 17 | Tspan13 | 1953 | 0.348 | 0.1374 | No |
| 18 | Evi5 | 2236 | 0.314 | 0.1250 | No |
| 19 | Vwa5a | 2315 | 0.304 | 0.1288 | No |
| 20 | Akt2 | 2338 | 0.301 | 0.1372 | No |
| 21 | Itgb2 | 2401 | 0.294 | 0.1421 | No |
| 22 | Cd37 | 2436 | 0.289 | 0.1490 | No |
| 23 | Hdac9 | 2480 | 0.283 | 0.1550 | No |
| 24 | Ammecr1 | 2592 | 0.272 | 0.1552 | No |
| 25 | Tnfrsf1b | 3017 | 0.232 | 0.1284 | No |
| 26 | Tmem158 | 3141 | 0.224 | 0.1259 | No |
| 27 | Tfpi | 3234 | 0.216 | 0.1257 | No |
| 28 | Il1rl2 | 3308 | 0.209 | 0.1268 | No |
| 29 | Igf2 | 3380 | 0.205 | 0.1279 | No |
| 30 | Plaur | 3465 | 0.200 | 0.1278 | No |
| 31 | Fbxo4 | 3713 | 0.179 | 0.1137 | No |
| 32 | Map4k1 | 3912 | 0.162 | 0.1030 | No |
| 33 | Ank | 4051 | 0.151 | 0.0968 | No |
| 34 | Traf1 | 4159 | 0.142 | 0.0929 | No |
| 35 | Zfp277 | 4267 | 0.134 | 0.0887 | No |
| 36 | Abcb1a | 4369 | 0.127 | 0.0847 | No |
| 37 | Jup | 4383 | 0.125 | 0.0879 | No |
| 38 | Epb41l3 | 4643 | 0.104 | 0.0702 | No |
| 39 | Pdcd1lg2 | 4669 | 0.101 | 0.0716 | No |
| 40 | Crot | 4721 | 0.097 | 0.0707 | No |
| 41 | Rbm4 | 4756 | 0.094 | 0.0711 | No |
| 42 | Laptm5 | 4878 | 0.085 | 0.0641 | No |
| 43 | Cab39l | 4891 | 0.085 | 0.0660 | No |
| 44 | Eng | 5207 | 0.061 | 0.0423 | No |
| 45 | Gng11 | 5356 | 0.051 | 0.0320 | No |
| 46 | Adgra2 | 5758 | 0.024 | 0.0001 | No |
| 47 | Pecam1 | 5901 | 0.012 | -0.0111 | No |
| 48 | St6gal1 | 6174 | -0.002 | -0.0332 | No |
| 49 | Ccser2 | 6272 | -0.009 | -0.0408 | No |
| 50 | Dcbld2 | 6277 | -0.009 | -0.0409 | No |
| 51 | Spry2 | 6520 | -0.026 | -0.0597 | No |
| 52 | Cbr4 | 6585 | -0.031 | -0.0639 | No |
| 53 | Inhba | 6598 | -0.032 | -0.0638 | No |
| 54 | Csf2ra | 6642 | -0.035 | -0.0661 | No |
| 55 | Peg3 | 6706 | -0.039 | -0.0700 | No |
| 56 | Tmem176b | 6951 | -0.056 | -0.0880 | No |
| 57 | Gucy1a1 | 7010 | -0.059 | -0.0907 | No |
| 58 | Lcp1 | 7208 | -0.071 | -0.1044 | No |
| 59 | Satb1 | 7261 | -0.075 | -0.1061 | No |
| 60 | Fuca1 | 7408 | -0.083 | -0.1152 | No |
| 61 | Trib2 | 7423 | -0.084 | -0.1135 | No |
| 62 | Etv5 | 7476 | -0.088 | -0.1148 | No |
| 63 | Il10ra | 7736 | -0.107 | -0.1323 | No |
| 64 | Mtmr10 | 7835 | -0.114 | -0.1365 | No |
| 65 | Map3k1 | 8135 | -0.133 | -0.1564 | No |
| 66 | Ero1a | 8320 | -0.146 | -0.1665 | No |
| 67 | Rabgap1l | 8366 | -0.149 | -0.1651 | No |
| 68 | Map7 | 8539 | -0.160 | -0.1737 | No |
| 69 | Mycn | 8661 | -0.169 | -0.1779 | No |
| 70 | Tnfaip3 | 8893 | -0.188 | -0.1904 | No |
| 71 | Avl9 | 9040 | -0.198 | -0.1957 | No |
| 72 | Klf4 | 9170 | -0.209 | -0.1991 | No |
| 73 | Ptcd2 | 9295 | -0.217 | -0.2020 | No |
| 74 | Cdadc1 | 9612 | -0.245 | -0.2195 | Yes |
| 75 | Itga2 | 9708 | -0.253 | -0.2187 | Yes |
| 76 | Cxcr4 | 9717 | -0.253 | -0.2108 | Yes |
| 77 | Ctss | 9790 | -0.260 | -0.2079 | Yes |
| 78 | Gadd45g | 9813 | -0.261 | -0.2009 | Yes |
| 79 | F13a1 | 9859 | -0.265 | -0.1957 | Yes |
| 80 | Ikzf1 | 9977 | -0.275 | -0.1959 | Yes |
| 81 | Atg10 | 9989 | -0.276 | -0.1875 | Yes |
| 82 | Sdccag8 | 9992 | -0.277 | -0.1784 | Yes |
| 83 | Irf8 | 10249 | -0.301 | -0.1891 | Yes |
| 84 | Mmd | 10428 | -0.316 | -0.1930 | Yes |
| 85 | Prelid3b | 10528 | -0.328 | -0.1900 | Yes |
| 86 | Gypc | 10571 | -0.333 | -0.1822 | Yes |
| 87 | Btbd3 | 10723 | -0.349 | -0.1827 | Yes |
| 88 | Dusp6 | 10778 | -0.355 | -0.1752 | Yes |
| 89 | Adgrl4 | 10897 | -0.369 | -0.1724 | Yes |
| 90 | Cbl | 11090 | -0.399 | -0.1746 | Yes |
| 91 | Tmem176a | 11109 | -0.403 | -0.1625 | Yes |
| 92 | Zfp639 | 11152 | -0.410 | -0.1521 | Yes |
| 93 | Gfpt2 | 11180 | -0.414 | -0.1403 | Yes |
| 94 | Ptbp2 | 11400 | -0.450 | -0.1430 | Yes |
| 95 | Wdr33 | 11506 | -0.469 | -0.1357 | Yes |
| 96 | Gprc5b | 11528 | -0.473 | -0.1215 | Yes |
| 97 | Psmb8 | 11532 | -0.474 | -0.1057 | Yes |
| 98 | Btc | 11558 | -0.480 | -0.0916 | Yes |
| 99 | Bpgm | 11639 | -0.495 | -0.0814 | Yes |
| 100 | Ccnd2 | 11816 | -0.535 | -0.0777 | Yes |
| 101 | Dock2 | 11853 | -0.546 | -0.0622 | Yes |
| 102 | Lat2 | 11869 | -0.549 | -0.0449 | Yes |
| 103 | Glrx | 11880 | -0.552 | -0.0271 | Yes |
| 104 | Yrdc | 12000 | -0.593 | -0.0169 | Yes |
| 105 | Car2 | 12048 | -0.612 | -0.0000 | Yes |
| 106 | Nin | 12246 | -0.761 | 0.0095 | Yes |