Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
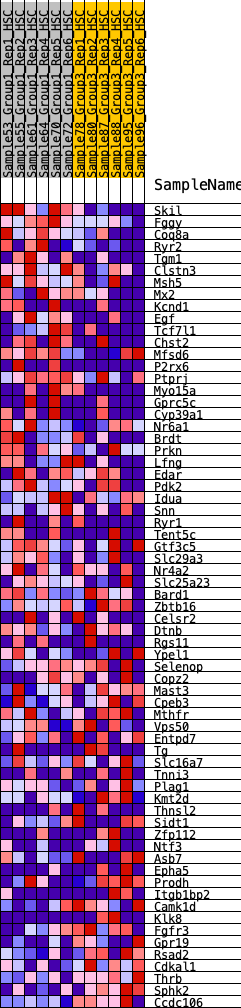
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
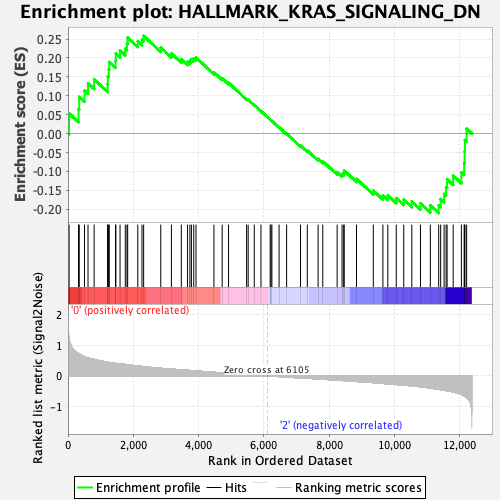
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
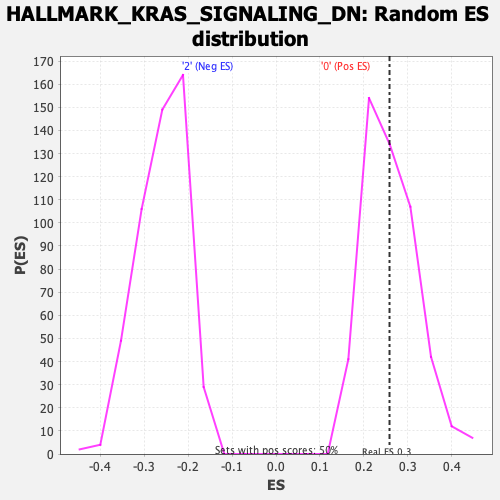
| Enrichment Score (ES) | 0.2583787 |
| Normalized Enrichment Score (NES) | 0.9880001 |
| Nominal p-value | 0.4607646 |
| FDR q-value | 0.8595524 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Skil | 33 | 1.193 | 0.0536 | Yes |
| 2 | Fggy | 323 | 0.736 | 0.0648 | Yes |
| 3 | Coq8a | 344 | 0.723 | 0.0972 | Yes |
| 4 | Ryr2 | 505 | 0.628 | 0.1138 | Yes |
| 5 | Tgm1 | 613 | 0.590 | 0.1329 | Yes |
| 6 | Clstn3 | 800 | 0.541 | 0.1433 | Yes |
| 7 | Msh5 | 1212 | 0.446 | 0.1309 | Yes |
| 8 | Mx2 | 1222 | 0.445 | 0.1512 | Yes |
| 9 | Kcnd1 | 1246 | 0.440 | 0.1700 | Yes |
| 10 | Egf | 1263 | 0.435 | 0.1893 | Yes |
| 11 | Tcf7l1 | 1455 | 0.412 | 0.1931 | Yes |
| 12 | Chst2 | 1465 | 0.410 | 0.2118 | Yes |
| 13 | Mfsd6 | 1593 | 0.395 | 0.2200 | Yes |
| 14 | P2rx6 | 1754 | 0.375 | 0.2247 | Yes |
| 15 | Ptprj | 1806 | 0.370 | 0.2380 | Yes |
| 16 | Myo15a | 1827 | 0.366 | 0.2537 | Yes |
| 17 | Gprc5c | 2136 | 0.323 | 0.2439 | Yes |
| 18 | Cyp39a1 | 2265 | 0.310 | 0.2481 | Yes |
| 19 | Nr6a1 | 2316 | 0.304 | 0.2584 | Yes |
| 20 | Brdt | 2840 | 0.249 | 0.2276 | No |
| 21 | Prkn | 3166 | 0.223 | 0.2117 | No |
| 22 | Lfng | 3469 | 0.199 | 0.1965 | No |
| 23 | Edar | 3660 | 0.185 | 0.1897 | No |
| 24 | Pdk2 | 3732 | 0.178 | 0.1923 | No |
| 25 | Idua | 3778 | 0.174 | 0.1969 | No |
| 26 | Snn | 3850 | 0.167 | 0.1990 | No |
| 27 | Ryr1 | 3919 | 0.162 | 0.2011 | No |
| 28 | Tent5c | 4468 | 0.118 | 0.1621 | No |
| 29 | Gtf3c5 | 4720 | 0.097 | 0.1462 | No |
| 30 | Slc29a3 | 4915 | 0.083 | 0.1344 | No |
| 31 | Nr4a2 | 5468 | 0.043 | 0.0915 | No |
| 32 | Slc25a23 | 5516 | 0.040 | 0.0896 | No |
| 33 | Bard1 | 5703 | 0.027 | 0.0757 | No |
| 34 | Zbtb16 | 5908 | 0.012 | 0.0597 | No |
| 35 | Celsr2 | 6190 | -0.003 | 0.0370 | No |
| 36 | Dtnb | 6214 | -0.005 | 0.0354 | No |
| 37 | Rgs11 | 6242 | -0.007 | 0.0335 | No |
| 38 | Ypel1 | 6462 | -0.023 | 0.0168 | No |
| 39 | Selenop | 6693 | -0.039 | -0.0001 | No |
| 40 | Copz2 | 7119 | -0.066 | -0.0315 | No |
| 41 | Mast3 | 7326 | -0.079 | -0.0446 | No |
| 42 | Cpeb3 | 7657 | -0.102 | -0.0666 | No |
| 43 | Mthfr | 7800 | -0.112 | -0.0729 | No |
| 44 | Vps50 | 8240 | -0.141 | -0.1020 | No |
| 45 | Entpd7 | 8390 | -0.151 | -0.1070 | No |
| 46 | Tg | 8442 | -0.154 | -0.1039 | No |
| 47 | Slc16a7 | 8455 | -0.155 | -0.0975 | No |
| 48 | Tnni3 | 8833 | -0.183 | -0.1196 | No |
| 49 | Plag1 | 9347 | -0.222 | -0.1508 | No |
| 50 | Kmt2d | 9641 | -0.247 | -0.1630 | No |
| 51 | Thnsl2 | 9791 | -0.260 | -0.1629 | No |
| 52 | Sidt1 | 10050 | -0.282 | -0.1706 | No |
| 53 | Zfp112 | 10279 | -0.303 | -0.1748 | No |
| 54 | Ntf3 | 10526 | -0.328 | -0.1794 | No |
| 55 | Asb7 | 10791 | -0.357 | -0.1840 | No |
| 56 | Epha5 | 11093 | -0.401 | -0.1896 | No |
| 57 | Prodh | 11349 | -0.440 | -0.1896 | No |
| 58 | Itgb1bp2 | 11409 | -0.452 | -0.1731 | No |
| 59 | Camk1d | 11515 | -0.471 | -0.1594 | No |
| 60 | Klk8 | 11576 | -0.484 | -0.1415 | No |
| 61 | Fgfr3 | 11606 | -0.489 | -0.1208 | No |
| 62 | Gpr19 | 11793 | -0.529 | -0.1110 | No |
| 63 | Rsad2 | 12044 | -0.611 | -0.1025 | No |
| 64 | Cdkal1 | 12128 | -0.661 | -0.0781 | No |
| 65 | Thrb | 12143 | -0.666 | -0.0478 | No |
| 66 | Sphk2 | 12152 | -0.671 | -0.0168 | No |
| 67 | Ccdc106 | 12204 | -0.719 | 0.0129 | No |