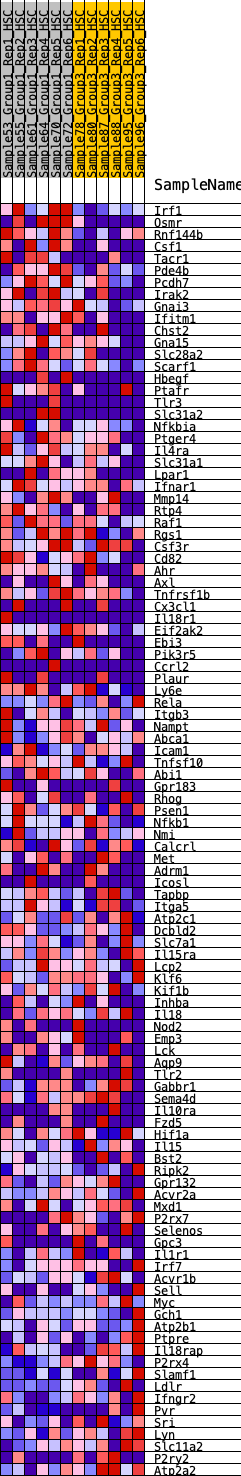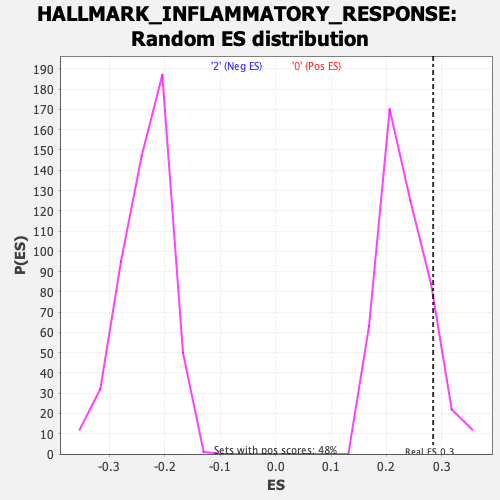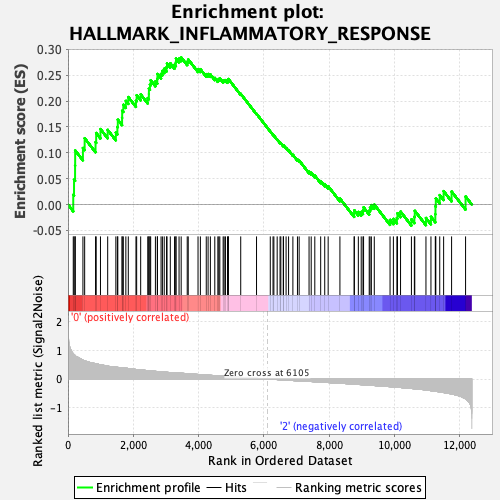
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.28444344 |
| Normalized Enrichment Score (NES) | 1.2250713 |
| Nominal p-value | 0.10714286 |
| FDR q-value | 0.6459255 |
| FWER p-Value | 0.937 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Irf1 | 161 | 0.874 | 0.0190 | Yes |
| 2 | Osmr | 185 | 0.844 | 0.0482 | Yes |
| 3 | Rnf144b | 217 | 0.807 | 0.0754 | Yes |
| 4 | Csf1 | 220 | 0.799 | 0.1046 | Yes |
| 5 | Tacr1 | 457 | 0.653 | 0.1094 | Yes |
| 6 | Pde4b | 510 | 0.625 | 0.1282 | Yes |
| 7 | Pcdh7 | 843 | 0.530 | 0.1206 | Yes |
| 8 | Irak2 | 863 | 0.525 | 0.1384 | Yes |
| 9 | Gnai3 | 994 | 0.494 | 0.1459 | Yes |
| 10 | Ifitm1 | 1215 | 0.446 | 0.1444 | Yes |
| 11 | Chst2 | 1465 | 0.410 | 0.1392 | Yes |
| 12 | Gna15 | 1512 | 0.403 | 0.1503 | Yes |
| 13 | Slc28a2 | 1525 | 0.402 | 0.1641 | Yes |
| 14 | Scarf1 | 1656 | 0.385 | 0.1676 | Yes |
| 15 | Hbegf | 1659 | 0.385 | 0.1816 | Yes |
| 16 | Ptafr | 1694 | 0.381 | 0.1929 | Yes |
| 17 | Tlr3 | 1771 | 0.375 | 0.2005 | Yes |
| 18 | Slc31a2 | 1846 | 0.364 | 0.2078 | Yes |
| 19 | Nfkbia | 2081 | 0.329 | 0.2009 | Yes |
| 20 | Ptger4 | 2102 | 0.327 | 0.2113 | Yes |
| 21 | Il4ra | 2225 | 0.315 | 0.2129 | Yes |
| 22 | Slc31a1 | 2442 | 0.288 | 0.2059 | Yes |
| 23 | Lpar1 | 2476 | 0.283 | 0.2136 | Yes |
| 24 | Ifnar1 | 2477 | 0.283 | 0.2241 | Yes |
| 25 | Mmp14 | 2509 | 0.279 | 0.2318 | Yes |
| 26 | Rtp4 | 2531 | 0.277 | 0.2403 | Yes |
| 27 | Raf1 | 2676 | 0.265 | 0.2383 | Yes |
| 28 | Rgs1 | 2734 | 0.261 | 0.2432 | Yes |
| 29 | Csf3r | 2739 | 0.260 | 0.2525 | Yes |
| 30 | Cd82 | 2849 | 0.248 | 0.2527 | Yes |
| 31 | Ahr | 2890 | 0.244 | 0.2584 | Yes |
| 32 | Axl | 2946 | 0.239 | 0.2627 | Yes |
| 33 | Tnfrsf1b | 3017 | 0.232 | 0.2656 | Yes |
| 34 | Cx3cl1 | 3031 | 0.231 | 0.2730 | Yes |
| 35 | Il18r1 | 3132 | 0.224 | 0.2731 | Yes |
| 36 | Eif2ak2 | 3262 | 0.213 | 0.2704 | Yes |
| 37 | Ebi3 | 3302 | 0.209 | 0.2749 | Yes |
| 38 | Pik3r5 | 3306 | 0.209 | 0.2824 | Yes |
| 39 | Ccrl2 | 3400 | 0.204 | 0.2823 | Yes |
| 40 | Plaur | 3465 | 0.200 | 0.2844 | Yes |
| 41 | Ly6e | 3650 | 0.186 | 0.2763 | No |
| 42 | Rela | 3681 | 0.182 | 0.2805 | No |
| 43 | Itgb3 | 3981 | 0.157 | 0.2619 | No |
| 44 | Nampt | 4053 | 0.150 | 0.2616 | No |
| 45 | Abca1 | 4233 | 0.136 | 0.2520 | No |
| 46 | Icam1 | 4289 | 0.132 | 0.2524 | No |
| 47 | Tnfsf10 | 4359 | 0.127 | 0.2515 | No |
| 48 | Abi1 | 4494 | 0.116 | 0.2448 | No |
| 49 | Gpr183 | 4588 | 0.107 | 0.2412 | No |
| 50 | Rhog | 4617 | 0.105 | 0.2428 | No |
| 51 | Psen1 | 4648 | 0.103 | 0.2441 | No |
| 52 | Nfkb1 | 4750 | 0.095 | 0.2393 | No |
| 53 | Nmi | 4780 | 0.093 | 0.2404 | No |
| 54 | Calcrl | 4820 | 0.090 | 0.2405 | No |
| 55 | Met | 4884 | 0.085 | 0.2385 | No |
| 56 | Adrm1 | 4904 | 0.084 | 0.2401 | No |
| 57 | Icosl | 4910 | 0.084 | 0.2427 | No |
| 58 | Tapbp | 5288 | 0.056 | 0.2140 | No |
| 59 | Itga5 | 5772 | 0.023 | 0.1754 | No |
| 60 | Atp2c1 | 6193 | -0.003 | 0.1413 | No |
| 61 | Dcbld2 | 6277 | -0.009 | 0.1349 | No |
| 62 | Slc7a1 | 6303 | -0.011 | 0.1333 | No |
| 63 | Il15ra | 6404 | -0.018 | 0.1258 | No |
| 64 | Lcp2 | 6492 | -0.025 | 0.1196 | No |
| 65 | Klf6 | 6516 | -0.026 | 0.1187 | No |
| 66 | Kif1b | 6590 | -0.031 | 0.1139 | No |
| 67 | Inhba | 6598 | -0.032 | 0.1145 | No |
| 68 | Il18 | 6677 | -0.038 | 0.1095 | No |
| 69 | Nod2 | 6753 | -0.042 | 0.1049 | No |
| 70 | Emp3 | 6885 | -0.052 | 0.0961 | No |
| 71 | Lck | 7025 | -0.061 | 0.0870 | No |
| 72 | Aqp9 | 7077 | -0.064 | 0.0852 | No |
| 73 | Tlr2 | 7380 | -0.082 | 0.0636 | No |
| 74 | Gabbr1 | 7446 | -0.086 | 0.0614 | No |
| 75 | Sema4d | 7556 | -0.094 | 0.0560 | No |
| 76 | Il10ra | 7736 | -0.107 | 0.0453 | No |
| 77 | Fzd5 | 7860 | -0.115 | 0.0395 | No |
| 78 | Hif1a | 7965 | -0.121 | 0.0355 | No |
| 79 | Il15 | 8325 | -0.147 | 0.0116 | No |
| 80 | Bst2 | 8761 | -0.177 | -0.0174 | No |
| 81 | Ripk2 | 8766 | -0.177 | -0.0112 | No |
| 82 | Gpr132 | 8889 | -0.187 | -0.0142 | No |
| 83 | Acvr2a | 8977 | -0.194 | -0.0142 | No |
| 84 | Mxd1 | 9031 | -0.197 | -0.0113 | No |
| 85 | P2rx7 | 9049 | -0.199 | -0.0053 | No |
| 86 | Selenos | 9222 | -0.211 | -0.0116 | No |
| 87 | Gpc3 | 9247 | -0.213 | -0.0057 | No |
| 88 | Il1r1 | 9288 | -0.216 | -0.0010 | No |
| 89 | Irf7 | 9377 | -0.224 | 0.0000 | No |
| 90 | Acvr1b | 9860 | -0.265 | -0.0296 | No |
| 91 | Sell | 9962 | -0.274 | -0.0277 | No |
| 92 | Myc | 10070 | -0.284 | -0.0260 | No |
| 93 | Gch1 | 10087 | -0.285 | -0.0168 | No |
| 94 | Atp2b1 | 10181 | -0.294 | -0.0136 | No |
| 95 | Ptpre | 10514 | -0.326 | -0.0286 | No |
| 96 | Il18rap | 10604 | -0.336 | -0.0235 | No |
| 97 | P2rx4 | 10617 | -0.338 | -0.0121 | No |
| 98 | Slamf1 | 10959 | -0.380 | -0.0259 | No |
| 99 | Ldlr | 11112 | -0.403 | -0.0235 | No |
| 100 | Ifngr2 | 11246 | -0.425 | -0.0187 | No |
| 101 | Pvr | 11250 | -0.425 | -0.0033 | No |
| 102 | Sri | 11261 | -0.427 | 0.0116 | No |
| 103 | Lyn | 11381 | -0.446 | 0.0183 | No |
| 104 | Slc11a2 | 11500 | -0.469 | 0.0259 | No |
| 105 | P2ry2 | 11744 | -0.517 | 0.0251 | No |
| 106 | Atp2a2 | 12172 | -0.687 | 0.0156 | No |